
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,108,481 – 9,108,579 |
| Length | 98 |
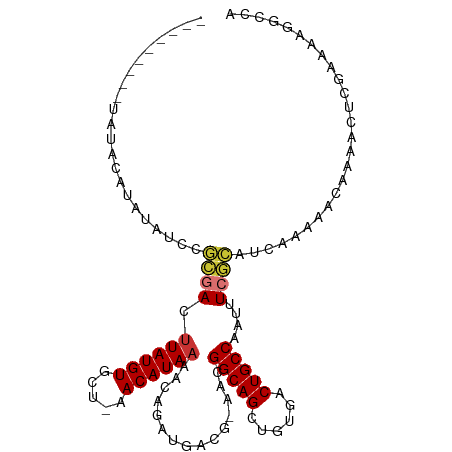
| Max. P | 0.987222 |

| Location | 9,108,481 – 9,108,579 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.20 |
| Shannon entropy | 0.46175 |
| G+C content | 0.43332 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.29 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
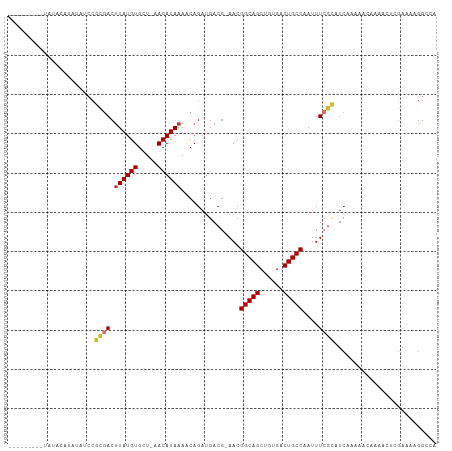
>dm3.chrX 9108481 98 + 22422827 ---------UAUACAUAUAUCCGCGACUUAUGUGCU-AACAUAAAACAGAUGACG-AACGGCAGCUGUGACUGCCAAUUUCGCAUCAAAAACAAAACUCGAAAAGGCCA ---------...........((.(((.((((((...-.))))))....((((.((-((.(((((......)))))...))))))))...........)))....))... ( -22.80, z-score = -2.80, R) >droEre2.scaffold_4690 12740792 82 - 18748788 ------------------------AACCUAUGUGCU-AACAUAAAGCAGAUGACG-AACGGCAGCUGUGACUGCCAAUUUCG-AUCCAAAACCAAACUCGAAUAGGCCA ------------------------..(((((.((((-.......)))).....((-((.(((((......)))))...))))-..................)))))... ( -17.80, z-score = -1.57, R) >droYak2.chrX 17705577 98 + 21770863 ---------CCAAUAUAUAUCCGCGACUUAUGUGCU-AACAUAAAACAGAUGACG-AACGGCAGCUGUGACUGCCAAUUUCGCAUCAAAAACAAAACUCGAAAUGGCCA ---------...........((.(((.((((((...-.))))))....((((.((-((.(((((......)))))...))))))))...........)))....))... ( -21.70, z-score = -2.22, R) >droSec1.super_345 10109 98 + 13804 ---------UAUACAUAUAUCCGCGACUUAUGUGCU-AACAUAAAACAGAUGACG-AACGGCAGCUGUGACUGCCAAUUUCGCAUCAAAAACAAAACUCGAAAAGGCCA ---------...........((.(((.((((((...-.))))))....((((.((-((.(((((......)))))...))))))))...........)))....))... ( -22.80, z-score = -2.80, R) >droSim1.chrX 7265760 98 + 17042790 ---------UAUACAUAUAUCCGCGACUUAUGUGCU-AACAUAAAACAGAUGACG-AACGGCAGCUGUGACUGCCAAUUUCGCAUCAAAAACAAAACUCGAAAAGGCCA ---------...........((.(((.((((((...-.))))))....((((.((-((.(((((......)))))...))))))))...........)))....))... ( -22.80, z-score = -2.80, R) >dp4.chrXL_group1a 2174777 104 - 9151740 CGAACACUCGGAACGCAGAUCCGCGACUUAUGUGUU-AACAUAAAACAGAUGACG-AACGGCAGCUGCGACUGCCAAUUUCGCUUCAAAACUUUGAGUGGAGGAGG--- ....((((((((.(((......)))..((((((...-.))))))....((.(.((-((.(((((......)))))...))))).)).....)))))))).......--- ( -28.20, z-score = -1.53, R) >droPer1.super_25 311988 104 + 1448063 CGAACACUCGGAACGCAGAUCCGCGACUUAUGUGUU-AACAUAAAACAGAUGACG-AACGGCAGCUGCGACUGCCAAUUUCGCUUCAAAACUUUGAGUGGAGGAGG--- ....((((((((.(((......)))..((((((...-.))))))....((.(.((-((.(((((......)))))...))))).)).....)))))))).......--- ( -28.20, z-score = -1.53, R) >droWil1.scaffold_181096 9897334 97 - 12416693 ---------UAUAGAUCUCCCCAUGACUUAUGUGUUUAACAUAAAACAGAUGACGGAACGGCAGCUGUGUCUGCCAAUUUGAUUUCAUCAACUUGGAUAUCCAUAG--- ---------(((.(((.(((...((..((((((.....))))))..))(((((((((..(((((......)))))..))))...))))).....))).))).))).--- ( -22.40, z-score = -1.60, R) >consensus _________UAUACAUAUAUCCGCGACUUAUGUGCU_AACAUAAAACAGAUGACG_AACGGCAGCUGUGACUGCCAAUUUCGCAUCAAAAACAAAACUCGAAAAGGCCA ......................((((.((((((.....))))))...............(((((......)))))....)))).......................... (-15.13 = -15.29 + 0.16)

| Location | 9,108,481 – 9,108,579 |
|---|---|
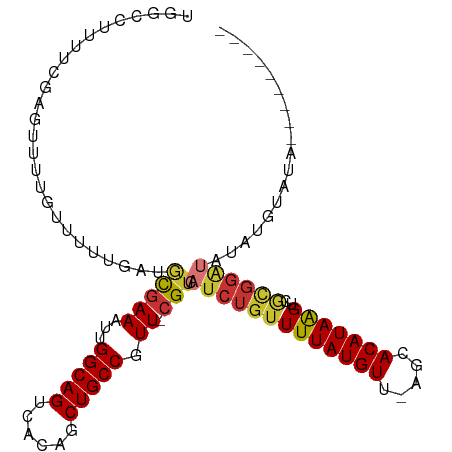
| Length | 98 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.20 |
| Shannon entropy | 0.46175 |
| G+C content | 0.43332 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
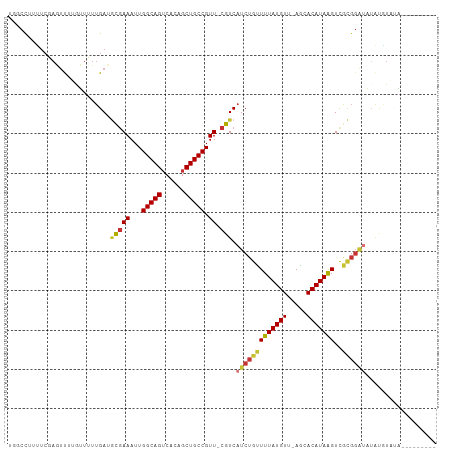
>dm3.chrX 9108481 98 - 22422827 UGGCCUUUUCGAGUUUUGUUUUUGAUGCGAAAUUGGCAGUCACAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AGCACAUAAGUCGCGGAUAUAUGUAUA--------- .(((.......((((((((.......))))))))((((((....))))))...-.)))(((((((((((((.-...)))))))..)))))).........--------- ( -25.30, z-score = -1.18, R) >droEre2.scaffold_4690 12740792 82 + 18748788 UGGCCUAUUCGAGUUUGGUUUUGGAU-CGAAAUUGGCAGUCACAGCUGCCGUU-CGUCAUCUGCUUUAUGUU-AGCACAUAGGUU------------------------ .(((((((.((((......))))((.-((((...((((((....)))))).))-))))...((((.......-)))).)))))))------------------------ ( -22.30, z-score = -1.09, R) >droYak2.chrX 17705577 98 - 21770863 UGGCCAUUUCGAGUUUUGUUUUUGAUGCGAAAUUGGCAGUCACAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AGCACAUAAGUCGCGGAUAUAUAUUGG--------- .(((.......((((((((.......))))))))((((((....))))))...-.)))(((((((((((((.-...)))))))..)))))).........--------- ( -25.30, z-score = -1.09, R) >droSec1.super_345 10109 98 - 13804 UGGCCUUUUCGAGUUUUGUUUUUGAUGCGAAAUUGGCAGUCACAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AGCACAUAAGUCGCGGAUAUAUGUAUA--------- .(((.......((((((((.......))))))))((((((....))))))...-.)))(((((((((((((.-...)))))))..)))))).........--------- ( -25.30, z-score = -1.18, R) >droSim1.chrX 7265760 98 - 17042790 UGGCCUUUUCGAGUUUUGUUUUUGAUGCGAAAUUGGCAGUCACAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AGCACAUAAGUCGCGGAUAUAUGUAUA--------- .(((.......((((((((.......))))))))((((((....))))))...-.)))(((((((((((((.-...)))))))..)))))).........--------- ( -25.30, z-score = -1.18, R) >dp4.chrXL_group1a 2174777 104 + 9151740 ---CCUCCUCCACUCAAAGUUUUGAAGCGAAAUUGGCAGUCGCAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AACACAUAAGUCGCGGAUCUGCGUUCCGAGUGUUCG ---.......(((((...........(((((...((((((....)))))).))-))).(((((((((((((.-...)))))))..)))))).........))))).... ( -29.20, z-score = -2.12, R) >droPer1.super_25 311988 104 - 1448063 ---CCUCCUCCACUCAAAGUUUUGAAGCGAAAUUGGCAGUCGCAGCUGCCGUU-CGUCAUCUGUUUUAUGUU-AACACAUAAGUCGCGGAUCUGCGUUCCGAGUGUUCG ---.......(((((...........(((((...((((((....)))))).))-))).(((((((((((((.-...)))))))..)))))).........))))).... ( -29.20, z-score = -2.12, R) >droWil1.scaffold_181096 9897334 97 + 12416693 ---CUAUGGAUAUCCAAGUUGAUGAAAUCAAAUUGGCAGACACAGCUGCCGUUCCGUCAUCUGUUUUAUGUUAAACACAUAAGUCAUGGGGAGAUCUAUA--------- ---.(((((((.(((.....(((((........((((((......)))))).....)))))((.(((((((.....))))))).))...))).)))))))--------- ( -27.12, z-score = -2.27, R) >consensus UGGCCUUUUCGAGUUUUGUUUUUGAUGCGAAAUUGGCAGUCACAGCUGCCGUU_CGUCAUCUGUUUUAUGUU_AGCACAUAAGUCGCGGAUAUAUGUAUA_________ ..........................(((.(((.(((((......)))))))).))).(((((((((((((.....)))))))..)))))).................. (-16.39 = -16.92 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:34 2011