
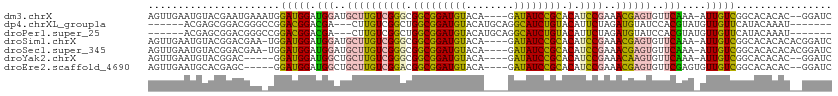
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,105,125 – 9,105,232 |
| Length | 107 |
| Max. P | 0.775702 |

| Location | 9,105,125 – 9,105,232 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.49344 |
| G+C content | 0.51690 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -20.13 |
| Energy contribution | -19.86 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9105125 107 + 22422827 AGUUGAAUGUACGAAUGAAAUGGAUGGAUGGAUGCUUGUCGGGCGGCGGAUGUACA----GAUAUCCGCACAUCCGAAACGAGUGUUCAAA-AUUGUCGGCACACAC--GGAUC ...........((..((...(.((..(.((((((((((((((..(((((((((...----.)))))))).)..))))..))))))))))..-.)..)).)..))..)--).... ( -31.40, z-score = -0.86, R) >dp4.chrXL_group1a 2170380 98 - 9151740 ------ACGAGCGGACGGGCCGGACGGACGA---CUUGUCGGCUGGCGGAUGUACAUGCAGGCAUCUGUACAUUCUAGAUGUAUCCACGUAUGUUGUUCAUACAAAU------- ------....(.(((...(((((.(.(((..---...))).))))))(((((((((((....))..)))))))))........))).)(((((.....)))))....------- ( -31.70, z-score = -0.98, R) >droPer1.super_25 307585 98 + 1448063 ------ACGAGCGGACGGGCCGGACGGACGA---CUUGUCGGCUGGCGGAUGUACAUGCAGGCAUCUGUACAUUCUAGAUGUAUCCACGUAUGUUGUUCAUACAAAU------- ------....(.(((...(((((.(.(((..---...))).))))))(((((((((((....))..)))))))))........))).)(((((.....)))))....------- ( -31.70, z-score = -0.98, R) >droSim1.chrX 7262399 108 + 17042790 AGUUGAAUGUACGGACGAA-UGGAUGGAUGGAUGCUUGUCGGGCGGCGGAUGUACA----GAUAUCCGCACAUCCGAAACGAGUGUUCAAA-AUUGUCGGCACACACACGGAUC .((.(..(((.(.(((((.-........((((((((((((((..(((((((((...----.)))))))).)..))))..))))))))))..-.))))).).)))..)))..... ( -33.50, z-score = -0.97, R) >droSec1.super_345 6814 108 + 13804 AGUUGAAUGUACGGACGAA-UGGAUGGAUGGAUGCUUGUCGGGCGGCGGAUGUACA----GAUAUCCGCACAUCCGAAACGAGUGUUCAAA-AUUGUCGGCACACACACGGAUC .((.(..(((.(.(((((.-........((((((((((((((..(((((((((...----.)))))))).)..))))..))))))))))..-.))))).).)))..)))..... ( -33.50, z-score = -0.97, R) >droYak2.chrX 17702181 102 + 21770863 AGUUGAAUGUACGGAC-----GGAUGGAUGGCUGCUUGUCGGGCGGCGGAUGUACA----GAUAUCCGCACAUCCGAAACAAGUGUUCAAA-AUUGUCGGCACACAC--GGAUC ..(((..(((.(.(((-----((.(((((....(((((((((..(((((((((...----.)))))))).)..))))..))))))))))..-.))))).).)))..)--))... ( -31.70, z-score = -0.88, R) >droEre2.scaffold_4690 12737576 103 - 18748788 AGUUGAAUGCACGAGC-----GGAUGGAUGGCUGCUUGUCGGACGGCGGAUGUACA----GAUAUCCGCACAUCCGAAACGAGUGUUCGAGUGUUGUCGGCACACAC--GGAUC ..((((((((.(((((-----((........)))))))(((((.(((((((((...----.)))))))).).))))).....))))))))((((((....)).))))--..... ( -39.50, z-score = -1.88, R) >consensus AGUUGAAUGUACGGACGAA_UGGAUGGAUGGAUGCUUGUCGGGCGGCGGAUGUACA____GAUAUCCGCACAUCCGAAACGAGUGUUCAAA_AUUGUCGGCACACAC__GGAUC ......................(((((.(((.(((((((((((.(((((((((........)))))))).).))))..))))))).)))....)))))................ (-20.13 = -19.86 + -0.27)


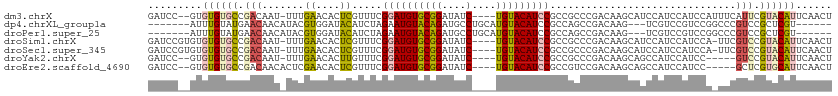
| Location | 9,105,125 – 9,105,232 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.49344 |
| G+C content | 0.51690 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9105125 107 - 22422827 GAUCC--GUGUGUGCCGACAAU-UUUGAACACUCGUUUCGGAUGUGCGGAUAUC----UGUACAUCCGCCGCCCGACAAGCAUCCAUCCAUCCAUUUCAUUCGUACAUUCAACU ((..(--(.((((..(((....-.))).)))).))..))((((((((((....)----)))))))))...((.......))................................. ( -26.50, z-score = -1.45, R) >dp4.chrXL_group1a 2170380 98 + 9151740 -------AUUUGUAUGAACAACAUACGUGGAUACAUCUAGAAUGUACAGAUGCCUGCAUGUACAUCCGCCAGCCGACAAG---UCGUCCGUCCGGCCCGUCCGCUCGU------ -------....(((((.....)))))((((((.......(.(((((((..((....))))))))).)(((.((.(((...---..))).))..)))..))))))....------ ( -27.10, z-score = -1.18, R) >droPer1.super_25 307585 98 - 1448063 -------AUUUGUAUGAACAACAUACGUGGAUACAUCUAGAAUGUACAGAUGCCUGCAUGUACAUCCGCCAGCCGACAAG---UCGUCCGUCCGGCCCGUCCGCUCGU------ -------....(((((.....)))))((((((.......(.(((((((..((....))))))))).)(((.((.(((...---..))).))..)))..))))))....------ ( -27.10, z-score = -1.18, R) >droSim1.chrX 7262399 108 - 17042790 GAUCCGUGUGUGUGCCGACAAU-UUUGAACACUCGUUUCGGAUGUGCGGAUAUC----UGUACAUCCGCCGCCCGACAAGCAUCCAUCCAUCCA-UUCGUCCGUACAUUCAACU .....((..((((((.(((...-...............(((((((((((....)----))))))))))..((.......)).............-...))).))))))...)). ( -30.60, z-score = -1.91, R) >droSec1.super_345 6814 108 - 13804 GAUCCGUGUGUGUGCCGACAAU-UUUGAACACUCGUUUCGGAUGUGCGGAUAUC----UGUACAUCCGCCGCCCGACAAGCAUCCAUCCAUCCA-UUCGUCCGUACAUUCAACU .....((..((((((.(((...-...............(((((((((((....)----))))))))))..((.......)).............-...))).))))))...)). ( -30.60, z-score = -1.91, R) >droYak2.chrX 17702181 102 - 21770863 GAUCC--GUGUGUGCCGACAAU-UUUGAACACUUGUUUCGGAUGUGCGGAUAUC----UGUACAUCCGCCGCCCGACAAGCAGCCAUCCAUCC-----GUCCGUACAUUCAACU .....--(.((((((.(((...-...((((....)))).((((((((((....)----)))))))))((.((.......)).)).........-----))).)))))).).... ( -30.90, z-score = -2.48, R) >droEre2.scaffold_4690 12737576 103 + 18748788 GAUCC--GUGUGUGCCGACAACACUCGAACACUCGUUUCGGAUGUGCGGAUAUC----UGUACAUCCGCCGUCCGACAAGCAGCCAUCCAUCC-----GCUCGUGCAUUCAACU ((..(--(.((((..(((......))).)))).))..))((((((((((....)----)))))))))((((..((.................)-----)..)).))........ ( -32.23, z-score = -1.79, R) >consensus GAUCC__GUGUGUGCCGACAAU_UUUGAACACUCGUUUCGGAUGUGCGGAUAUC____UGUACAUCCGCCGCCCGACAAGCAUCCAUCCAUCCA_UUCGUCCGUACAUUCAACU .........((((((.(((.......((....)).....(((((((((..........)))))))))...............................))).))))))...... (-14.14 = -14.31 + 0.17)
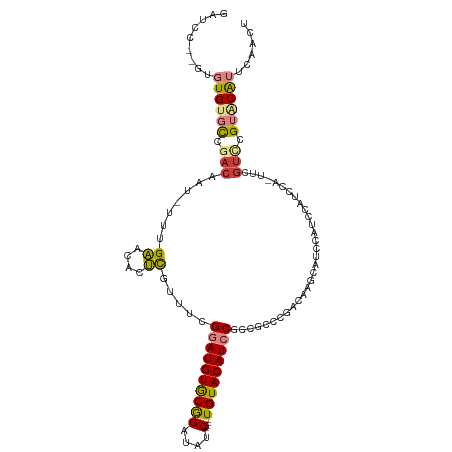

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:32 2011