
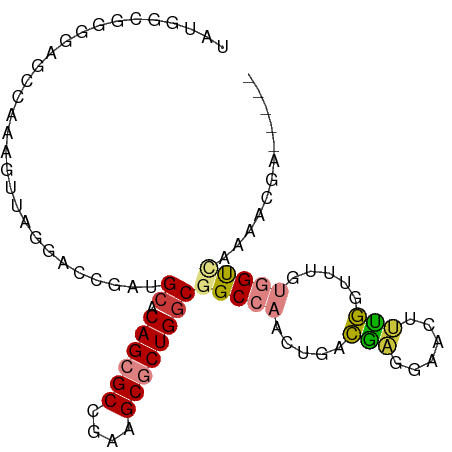
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,048,977 – 9,049,066 |
| Length | 89 |
| Max. P | 0.994677 |

| Location | 9,048,977 – 9,049,066 |
|---|---|
| Length | 89 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.53771 |
| G+C content | 0.60588 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 9048977 89 + 22422827 UAUCGCUGGGAGCCAAAGUUUGGACCGAUGCACAGCGCCGAAGCGCUGGCGGCCAACUGACGAGGAACUUUGGUUCGUGGUCAUAACGA----- (((.((((.((((((((((((((.(((.....((((((....)))))).)))))..((....)))))))))))))).)))).)))....----- ( -37.80, z-score = -3.06, R) >droEre2.scaffold_4690 12679740 89 - 18748788 UAUGGCGGGGAGCCAAAAUUAGGACCGAUGCACAGCGCCGAAGCGCUGGCGGCCAACUGACGAGGAACUUUGGUUUGGGGUCACAACGA----- ..((((.....)))).......((((..(((.((((((....))))))))).(((((..(.(.....).)..).)))))))).......----- ( -30.50, z-score = -1.34, R) >droYak2.chrX 9502769 89 - 21770863 UGUGGCGGGGAGCCAAAGUUAGGACCGAUGCACAGCGCCGAAGCGCUGGCGGCCAACUGACAAGGAACUUUGGUUCGUGGUUACAACGA----- ((((((.(.(((((((((((.((.(((.....((((((....)))))).)))))..((....)).))))))))))).).))))))....----- ( -41.70, z-score = -4.14, R) >droSec1.super_34 407812 89 + 457129 UAUGGCGGGGAGCCAAAGUUUGGACCGAUGCACAGCGCCGAAGCGCUGGCGGCCAACUGACGAGGAACUUUGGUUCGUCGUCACAACGA----- ..((((((.((((((((((((((.(((.....((((((....)))))).)))))..((....)))))))))))))).))))))......----- ( -42.80, z-score = -4.22, R) >dp4.chrXL_group1e 5591993 91 - 12523060 UAGAGCCCAGAGCCCAAAC---GGGCGAUGCUCAGCGCCGGAGCACUGGCAGCCAACUGUGGCCGAUGAGUCCGCUGUGGCCCAGAGGAAACCC ..((((.....((((....---))))...)))).(((((((....))))).))...(((.(((((...((....)).)))))))).(....).. ( -35.40, z-score = -1.12, R) >droPer1.super_25 1179967 91 - 1448063 UAGAGCCCAGAGCCCAAAC---GGGCGAUGCUCAGCGCCGGAGCACUGGCAGCCAACUGUGGCCGAUGAGUCCGCUGUGGCCCAGAGGAAACCC ..((((.....((((....---))))...)))).(((((((....))))).))...(((.(((((...((....)).)))))))).(....).. ( -35.40, z-score = -1.12, R) >droAna3.scaffold_13047 1387504 82 - 1816235 -------GGAAGCCAAAGUUAAGACCGAUGUGCAGCGCCGAAGCGCUGCCGGCCAACUGACGAAGCAUAGCGGCUUGGAGCCAAAACGA----- -------((...((...((((.(.(((....(((((((....)))))))))).)...)))).((((......))))))..)).......----- ( -25.20, z-score = -0.42, R) >consensus UAUGGCGGGGAGCCAAAGUUAGGACCGAUGCACAGCGCCGAAGCGCUGGCGGCCAACUGACGAGGAACUUUGGUUUGUGGUCAAAACGA_____ .............................((.((((((....))))))))(((((.....(((......))).....)))))............ (-15.52 = -15.64 + 0.13)

| Location | 9,048,977 – 9,049,066 |
|---|---|
| Length | 89 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.53771 |
| G+C content | 0.60588 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
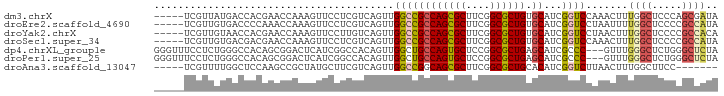
>dm3.chrX 9048977 89 - 22422827 -----UCGUUAUGACCACGAACCAAAGUUCCUCGUCAGUUGGCCGCCAGCGCUUCGGCGCUGUGCAUCGGUCCAAACUUUGGCUCCCAGCGAUA -----(((((.((((...((((....))))...))))(..((((((((((((....)))))).))...(((....)))..))))..)))))).. ( -33.60, z-score = -3.02, R) >droEre2.scaffold_4690 12679740 89 + 18748788 -----UCGUUGUGACCCCAAACCAAAGUUCCUCGUCAGUUGGCCGCCAGCGCUUCGGCGCUGUGCAUCGGUCCUAAUUUUGGCUCCCCGCCAUA -----..((((.((((((((......(.....).....))))..((((((((....)))))).))...)))).))))..((((.....)))).. ( -30.20, z-score = -2.88, R) >droYak2.chrX 9502769 89 + 21770863 -----UCGUUGUAACCACGAACCAAAGUUCCUUGUCAGUUGGCCGCCAGCGCUUCGGCGCUGUGCAUCGGUCCUAACUUUGGCUCCCCGCCACA -----..((((..(((..((((....))))...(((....))).((((((((....)))))).))...)))..))))..((((.....)))).. ( -31.40, z-score = -2.69, R) >droSec1.super_34 407812 89 - 457129 -----UCGUUGUGACGACGAACCAAAGUUCCUCGUCAGUUGGCCGCCAGCGCUUCGGCGCUGUGCAUCGGUCCAAACUUUGGCUCCCCGCCAUA -----......((((((.((((....)))).)))))).((((((((((((((....)))))).))...)).))))....((((.....)))).. ( -38.70, z-score = -4.20, R) >dp4.chrXL_group1e 5591993 91 + 12523060 GGGUUUCCUCUGGGCCACAGCGGACUCAUCGGCCACAGUUGGCUGCCAGUGCUCCGGCGCUGAGCAUCGCCC---GUUUGGGCUCUGGGCUCUA ((((..((...(((((..(((((.......(((((....)))))((((((((....)))))).)).....))---)))..))))).)))))).. ( -38.20, z-score = -0.30, R) >droPer1.super_25 1179967 91 + 1448063 GGGUUUCCUCUGGGCCACAGCGGACUCAUCGGCCACAGUUGGCUGCCAGUGCUCCGGCGCUGAGCAUCGCCC---GUUUGGGCUCUGGGCUCUA ((((..((...(((((..(((((.......(((((....)))))((((((((....)))))).)).....))---)))..))))).)))))).. ( -38.20, z-score = -0.30, R) >droAna3.scaffold_13047 1387504 82 + 1816235 -----UCGUUUUGGCUCCAAGCCGCUAUGCUUCGUCAGUUGGCCGGCAGCGCUUCGGCGCUGCACAUCGGUCUUAACUUUGGCUUCC------- -----.(((..((((.....))))..)))....(((((..((((((((((((....)))))))....)))))......)))))....------- ( -32.60, z-score = -2.75, R) >consensus _____UCGUUGUGACCACAAACCAAAGUUCCUCGUCAGUUGGCCGCCAGCGCUUCGGCGCUGUGCAUCGGUCCUAACUUUGGCUCCCCGCCAUA ........................................((((((((((((....)))))).))...)))).......((((.....)))).. (-18.94 = -19.03 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:30 2011