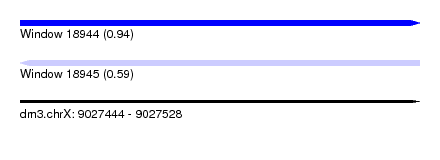
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,027,444 – 9,027,528 |
| Length | 84 |
| Max. P | 0.942529 |

| Location | 9,027,444 – 9,027,528 |
|---|---|
| Length | 84 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 65.03 |
| Shannon entropy | 0.56274 |
| G+C content | 0.35829 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -14.93 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942529 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
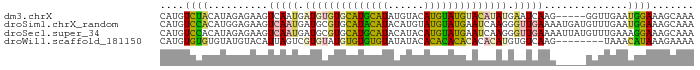
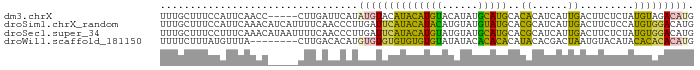
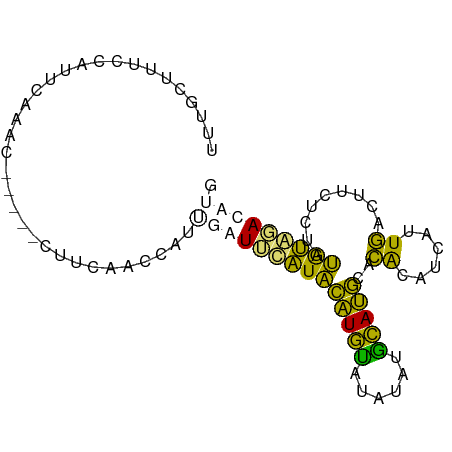
>dm3.chrX 9027444 84 + 22422827 CAUGUCUACAUAGAGAAGUCAAUGAUGUGUGCAUGCAUAUGUACAUGUAUGUACAUAUGAAUCAAG-----GGUUGAAUGGAAAGCAAA (((((.(((((...(....)....))))).)))))((((((((((....)))))))))).......-----.(((........)))... ( -19.20, z-score = -0.56, R) >droSim1.chrX_random 2659929 89 + 5698898 CAUGUCCACAUGGAGAAGUCAAUGAUGCGUGCAUACAUACAUGUAUGUAUGAAUCAAGGGUUGAAAAUGAUGUUUGAAUGGAAAGCAAA ((((....))))....(..(..((((.((((((((((....)))))))))).))))..)..)........(((((.......))))).. ( -21.00, z-score = -1.04, R) >droSec1.super_34 387634 89 + 457129 CAUGUCCACAUAGAGAAGUCAAUGAUGCGUGCAUGCAUACAUACAUGUAUGAAUCAAGGGUUGAAAAUUAUGUUUGAAAGGAAAGCAAA ....(((((((((...(..(..((((.((((((((........)))))))).))))..)..).....))))))......)))....... ( -17.70, z-score = -0.47, R) >droWil1.scaffold_181150 2031128 81 + 4952429 CAUGUGUGUGUAUGUACAUUAGUCGUGUAUGUGUGUGUAUAUACACACACACACACAUGUGUCAAG--------UAAACAUAAAGAAAA (((((((((((..((((((.....))))))((((((((....))))))))))))))))))).....--------............... ( -29.00, z-score = -2.12, R) >consensus CAUGUCCACAUAGAGAAGUCAAUGAUGCGUGCAUGCAUACAUACAUGUAUGAACAAAGGGUUCAAA_____GUUUGAAUGGAAAGCAAA ....((((((........(((((.((((((((((((((......)))))))))))))).)))))..........))..))))....... (-14.93 = -14.74 + -0.19)
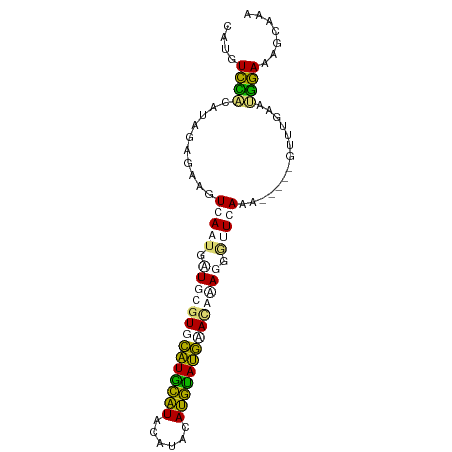
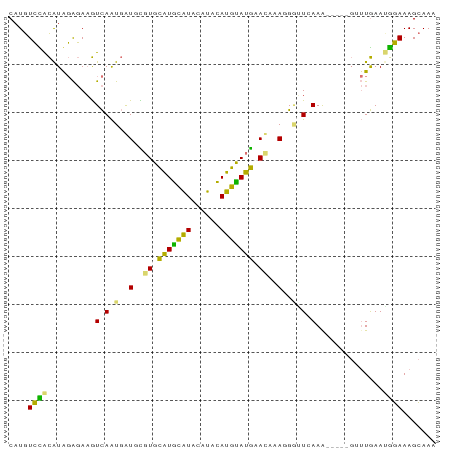
| Location | 9,027,444 – 9,027,528 |
|---|---|
| Length | 84 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 65.03 |
| Shannon entropy | 0.56274 |
| G+C content | 0.35829 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -8.70 |
| Energy contribution | -7.08 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.81 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586354 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 9027444 84 - 22422827 UUUGCUUUCCAUUCAACC-----CUUGAUUCAUAUGUACAUACAUGUACAUAUGCAUGCACACAUCAUUGACUUCUCUAUGUAGACAUG ..................-----.......((((((((((....))))))))))((((...((((....(....)...))))...)))) ( -12.70, z-score = -0.70, R) >droSim1.chrX_random 2659929 89 - 5698898 UUUGCUUUCCAUUCAAACAUCAUUUUCAACCCUUGAUUCAUACAUACAUGUAUGUAUGCACGCAUCAUUGACUUCUCCAUGUGGACAUG .......(((((.............((((....((...((((((((....))))))))....))...)))).........))))).... ( -12.95, z-score = -0.43, R) >droSec1.super_34 387634 89 - 457129 UUUGCUUUCCUUUCAAACAUAAUUUUCAACCCUUGAUUCAUACAUGUAUGUAUGCAUGCACGCAUCAUUGACUUCUCUAUGUGGACAUG ......................................((((((....))))))((((..(((((....(....)...)))))..)))) ( -11.20, z-score = 0.22, R) >droWil1.scaffold_181150 2031128 81 - 4952429 UUUUCUUUAUGUUUA--------CUUGACACAUGUGUGUGUGUGUGUAUAUACACACACAUACACGACUAAUGUACAUACACACACAUG .........((((..--------...))))(((((((((((((((((....)))))))).((((.......)))).....))))))))) ( -25.40, z-score = -2.24, R) >consensus UUUGCUUUCCAUUCAAAC_____CUUCAACCAUUGAUUCAUACAUGUAUAUAUGCAUGCACACAUCAUUGACUUCUCUAUGUAGACAUG .................................((((((((((((((......)))))..((......)).........))))))))). ( -8.70 = -7.08 + -1.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:28 2011