
| Sequence ID | dm3.chrX |
|---|---|
| Location | 9,025,239 – 9,025,354 |
| Length | 115 |
| Max. P | 0.982318 |

| Location | 9,025,239 – 9,025,354 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Shannon entropy | 0.12053 |
| G+C content | 0.51808 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
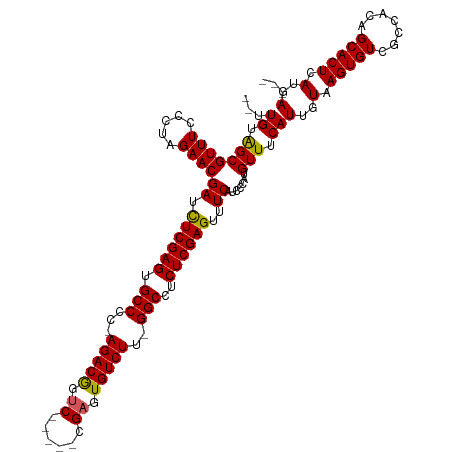
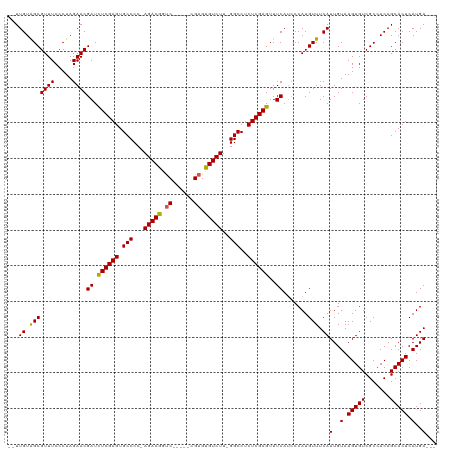
>dm3.chrX 9025239 115 + 22422827 --UUGUAGCGUUUCCCUAGAACGAUCUCGAGUGCCCCCAGACGGUCCGGUCCGAGUGUCUUUGGCCUCUCGAGUUUCAGACCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGA--- --.((.(((((((.....))))((.((((((.(((...(((((.((......)).)))))..)))..))))))..)).......))).))(..(.(((((.......))))).)..)--- ( -29.00, z-score = -0.01, R) >droSim1.chrX_random 2657829 108 + 5698898 --UUGUGGCGUUUCCCUAGAACGAUCUCGAGUGCCCC-AGACGGUC-----CGAGUGUCUU-GGCCUCUCGAGUUUCAUCCCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGA--- --(((((((((((.....))))((.((((((.(((..-(((((...-----....))))).-)))..))))))..))........................))))))).........--- ( -30.90, z-score = -1.21, R) >droSec1.super_34 385546 108 + 457129 --UUGUGGCGUUUCCCUAGAACGAUCUCGAGUGCCCC-AGACGGCC-----CGAGUGUCUU-GGCCUCUCGAGUUUCAUCCCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGA--- --(((((((((((.....))))((.((((((.(((..-(((((...-----....))))).-)))..))))))..))........................))))))).........--- ( -30.90, z-score = -1.22, R) >droYak2.chrX 9479747 113 - 21770863 CAUUGUAGCGUUUCCCUAGAACGAUUUCGAGUGCCCC-AGACAGUC-----CGAGUGUCUU-GGCCUCUCGAGUUUCAUCCCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGAGCA .......((((((.....))))((.((((((.(((..-(((((...-----....))))).-)))..))))))..)).............(..(.(((((.......))))).)..))). ( -25.00, z-score = -0.33, R) >droEre2.scaffold_4690 12655960 108 - 18748788 --UUGUAGCGUUUCCCUAGAACGAUUUCGAGUGCCCC-AGACAAUC-----CGAGUGUCUU-GGCCUCUCGAGUUUCAUCCCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGA--- --.((.(((((((.....))))((.((((((.(((..-(((((...-----....))))).-)))..))))))..)).......))).))(..(.(((((.......))))).)..)--- ( -23.60, z-score = -0.67, R) >consensus __UUGUAGCGUUUCCCUAGAACGAUCUCGAGUGCCCC_AGACGGUC_____CGAGUGUCUU_GGCCUCUCGAGUUUCAUCCCAUGUUUCAUUGUAAGUGUCGCCACAGCACUCAUGA___ ...((.(((((((.....))))((.((((((.(((...(((((.((......)).)))))..)))..))))))..)).......))).))(..(.(((((.......))))).)..)... (-27.56 = -27.04 + -0.52)

| Location | 9,025,239 – 9,025,354 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Shannon entropy | 0.12053 |
| G+C content | 0.51808 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -34.02 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
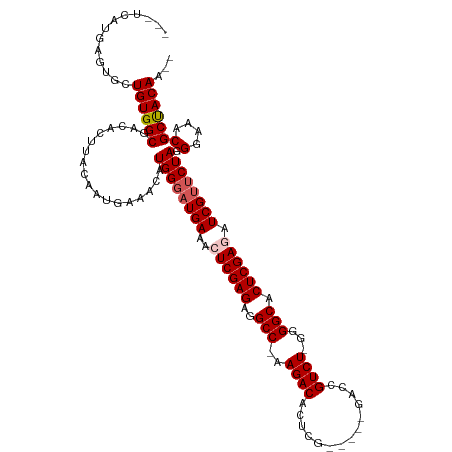
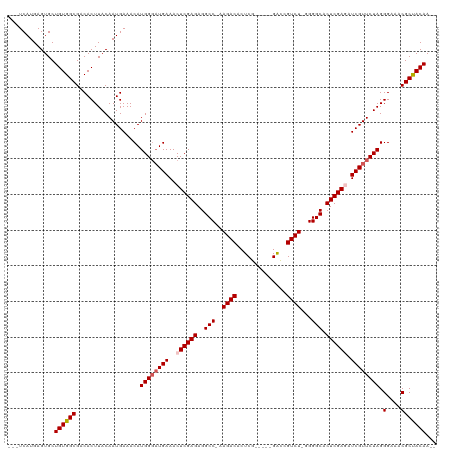
>dm3.chrX 9025239 115 - 22422827 ---UCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGUCUGAAACUCGAGAGGCCAAAGACACUCGGACCGGACCGUCUGGGGGCACUCGAGAUCGUUCUAGGGAAACGCUACAA-- ---..........(((((((...((((.(((((..((.....))...((((((..(((.......(((((((......)))))))))).)))))).))))).))))....))))))).-- ( -36.01, z-score = -1.13, R) >droSim1.chrX_random 2657829 108 - 5698898 ---UCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGGAUGAAACUCGAGAGGCC-AAGACACUCG-----GACCGUCU-GGGGCACUCGAGAUCGUUCUAGGGAAACGCCACAA-- ---..........((((((..................((((((((..((((((..(((-.((((.....-----....))))-..))).)))))).)))))))).(....))))))).-- ( -38.50, z-score = -2.88, R) >droSec1.super_34 385546 108 - 457129 ---UCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGGAUGAAACUCGAGAGGCC-AAGACACUCG-----GGCCGUCU-GGGGCACUCGAGAUCGUUCUAGGGAAACGCCACAA-- ---..........((((((..................((((((((..((((((..(((-.((((.....-----....))))-..))).)))))).)))))))).(....))))))).-- ( -38.50, z-score = -2.53, R) >droYak2.chrX 9479747 113 + 21770863 UGCUCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGGAUGAAACUCGAGAGGCC-AAGACACUCG-----GACUGUCU-GGGGCACUCGAAAUCGUUCUAGGGAAACGCUACAAUG .((........))((((((..................((((((((...(((((..(((-.(((((....-----...)))))-..))).)))))..)))))))).(....)))))))... ( -35.90, z-score = -2.60, R) >droEre2.scaffold_4690 12655960 108 + 18748788 ---UCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGGAUGAAACUCGAGAGGCC-AAGACACUCG-----GAUUGUCU-GGGGCACUCGAAAUCGUUCUAGGGAAACGCUACAA-- ---..........((((((..................((((((((...(((((..(((-.(((((....-----...)))))-..))).)))))..)))))))).(....))))))).-- ( -34.90, z-score = -2.85, R) >consensus ___UCAUGAGUGCUGUGGCGACACUUACAAUGAAACAUGGGAUGAAACUCGAGAGGCC_AAGACACUCG_____GACCGUCU_GGGGCACUCGAGAUCGUUCUAGGGAAACGCUACAA__ .............((((((..................((((((((..((((((..(((..((((..((......))..))))...))).)))))).)))))))).(....)))))))... (-34.02 = -34.42 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:26 2011