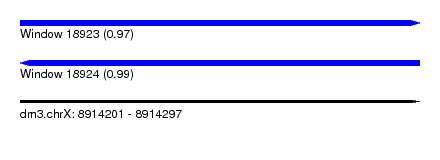
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,914,201 – 8,914,297 |
| Length | 96 |
| Max. P | 0.994894 |

| Location | 8,914,201 – 8,914,297 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.35 |
| Shannon entropy | 0.52487 |
| G+C content | 0.38958 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -10.56 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
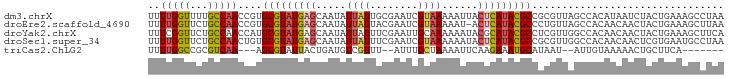
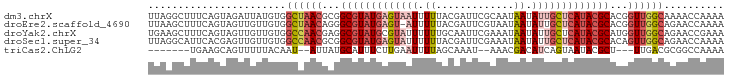
>dm3.chrX 8914201 96 + 22422827 UUUUGGUUUUGCCAACCGUGCGUAUGAGCAAUAUUAUUGCGAAUCGUAAAAAUUACUCAUACGCCGCGUUAGCCACAUAAUCUACUGAAAGCCUAA ....((((((((.(((((.(((((((((....(((.(((((...))))).)))..))))))))))).))).))..((........))))))))... ( -26.80, z-score = -3.25, R) >droEre2.scaffold_4690 12549327 95 - 18748788 UUUUGGUUCUGCCAACCGUGCGUAUGAGCAAUAUUAUUACGAAUCGUAAAAAU-ACUCAUACGCCCUGUUAGCCACAACAACUACUGAAAGCUUAA (((..((...((.(((.(.(((((((((...((((.(((((...))))).)))-)))))))))).).))).))..........))..)))...... ( -22.52, z-score = -2.93, R) >droYak2.chrX 9372229 96 - 21770863 UUUCGGUUCUGCCAACCAUGCGUAUGAGCAAUAUUAUUUCGAAUUGCAAAAAAUACGCAUACGCCUCGUUGGCCACAACAACUACUGAAAGCUUCA (((((((...((((((...(((((((.(((((..........)))))..........)))))))...))))))..........)))))))...... ( -23.82, z-score = -2.48, R) >droSec1.super_34 276925 96 + 457129 UUUUGGUUCUGCCAACUGUGCGUAUGAGCAAUAUUAUUUCGAAUCGUAAAAAAUACUCAUACGCCGCGUUGGCCACAACAACUCGUGAAUGCCUAA ....(((...((((((((.(((((((((...((((.(((((...)).))).))))))))))))))).))))))(((........)))...)))... ( -27.70, z-score = -3.14, R) >triCas2.ChLG2 316762 82 + 12900155 UUUUGGCCGCGUCAA---AGCGUAUUACUGAUGUCGUUU--AUUUGCUAAAAUUCAAGAAAUGCAUAAU--AUUGUAAAAACUGCUUCA------- ....((((((.....---.)))..((((.(((((..(.(--((((.((........))))))).)..))--))))))).....)))...------- ( -10.30, z-score = 1.21, R) >consensus UUUUGGUUCUGCCAACCGUGCGUAUGAGCAAUAUUAUUUCGAAUCGUAAAAAUUACUCAUACGCCGCGUUAGCCACAACAACUACUGAAAGCCUAA ..(((((...)))))....(((((((((.....((((........))))......)))))))))................................ (-10.56 = -10.92 + 0.36)
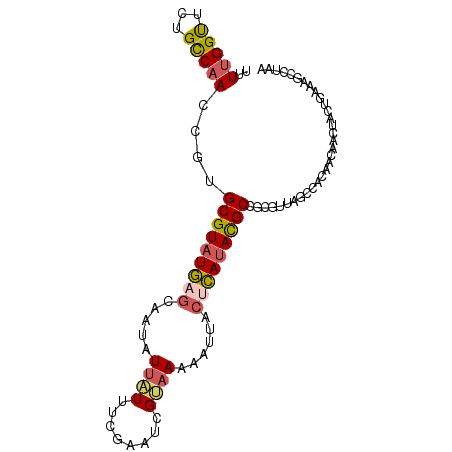
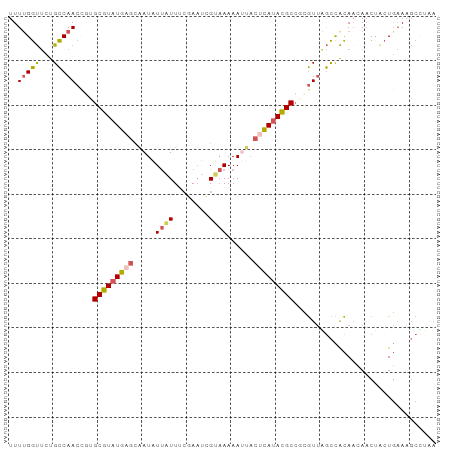
| Location | 8,914,201 – 8,914,297 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Shannon entropy | 0.52487 |
| G+C content | 0.38958 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -17.48 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8914201 96 - 22422827 UUAGGCUUUCAGUAGAUUAUGUGGCUAACGCGGCGUAUGAGUAAUUUUUACGAUUCGCAAUAAUAUUGCUCAUACGCACGGUUGGCAAAACCAAAA ...((((......))........((((((.(((((((((((((((...................))))))))))))).))))))))....)).... ( -29.11, z-score = -2.94, R) >droEre2.scaffold_4690 12549327 95 + 18748788 UUAAGCUUUCAGUAGUUGUUGUGGCUAACAGGGCGUAUGAGU-AUUUUUACGAUUCGUAAUAAUAUUGCUCAUACGCACGGUUGGCAGAACCAAAA ...((((......))))(((.((.(((((...((((((((((-(...(((((...)))))......)))))))))))...))))))).)))..... ( -31.20, z-score = -3.82, R) >droYak2.chrX 9372229 96 + 21770863 UGAAGCUUUCAGUAGUUGUUGUGGCCAACGAGGCGUAUGCGUAUUUUUUGCAAUUCGAAAUAAUAUUGCUCAUACGCAUGGUUGGCAGAACCGAAA ...((((......))))(((.((.(((((.(.(((((((.(((..(((((.....)))))......))).))))))).).))))))).)))..... ( -26.60, z-score = -1.14, R) >droSec1.super_34 276925 96 - 457129 UUAGGCAUUCACGAGUUGUUGUGGCCAACGCGGCGUAUGAGUAUUUUUUACGAUUCGAAAUAAUAUUGCUCAUACGCACAGUUGGCAGAACCAAAA ...((....(((((....)))))((((((...(((((((((((.......................)))))))))))...))))))....)).... ( -32.50, z-score = -3.87, R) >triCas2.ChLG2 316762 82 - 12900155 -------UGAAGCAGUUUUUACAAU--AUUAUGCAUUUCUUGAAUUUUAGCAAAU--AAACGACAUCAGUAAUACGCU---UUGACGCGGCCAAAA -------(((.(..((((......(--(..((.((.....)).))..))......--))))..).)))......(((.---.....)))....... ( -7.50, z-score = 1.78, R) >consensus UUAAGCUUUCAGUAGUUGUUGUGGCCAACGAGGCGUAUGAGUAAUUUUUACGAUUCGAAAUAAUAUUGCUCAUACGCACGGUUGGCAGAACCAAAA .......................((((((...(((((((((((((.((.............)).)))))))))))))...)))))).......... (-17.48 = -19.32 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:11 2011