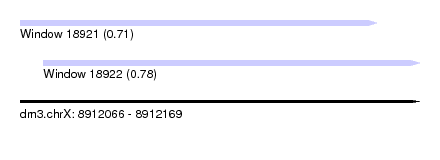
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,912,066 – 8,912,169 |
| Length | 103 |
| Max. P | 0.776690 |

| Location | 8,912,066 – 8,912,158 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Shannon entropy | 0.52612 |
| G+C content | 0.38962 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -9.53 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.705970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
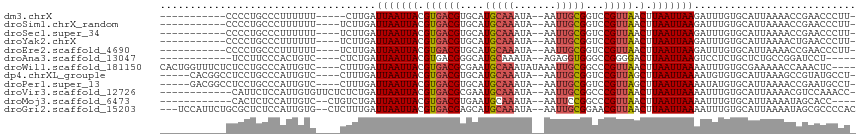
>dm3.chrX 8912066 92 + 22422827 ------------------CCACAUCC-CCUGCCCUUUUUU-----CUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAA ------------------........-..(((.(.....(-----(((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))..... ( -20.10, z-score = -2.94, R) >droSim1.chrX_random 2618595 90 + 5698898 ---------------------CAUCC-CCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAA ---------------------.....-..(((.(......----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))..... ( -19.50, z-score = -2.88, R) >droSec1.super_34 274779 93 + 457129 ------------------CCACAGCC-CCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAA ------------------.((((((.-...))........----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))..))))....... ( -20.40, z-score = -2.49, R) >droYak2.chrX 9370027 93 - 21770863 ------------------CCGCAGCC-CCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAA ------------------..((((..-.))))........----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))............. ( -20.80, z-score = -2.25, R) >droEre2.scaffold_4690 12547188 93 - 18748788 ------------------CCGCAGCC-CCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAA ------------------..((((..-.))))........----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))............. ( -20.80, z-score = -2.25, R) >droAna3.scaffold_13047 732380 88 + 1816235 ---------------------UGGGAUCCUUCCCACUGUC----CUCUGAUUAAUUACGUGACGGGCAUGCAAAUA--AGAGGUGGGCCGGGGACUUAAUUAAGUCCUCUGCUCU--- ---------------------(((((....)))))....(----((((.........((((.....))))......--))))).((((.((((((((....)))))))).)))).--- ( -30.96, z-score = -2.03, R) >droWil1.scaffold_181150 4196247 113 - 4952429 UUAUGUCGCUCGUUCACUGGUUUCUCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGCGAAUGCAAAUAUAAAUUGCGGCCCGUUAACUUAAUUAAAAUUUGUGCGAAAA- .....((((.((..((.(((............))).))..----)(((((((((.....(((((.(..(((((.......)))))..))))))..)))))))))....).))))...- ( -21.10, z-score = -0.76, R) >dp4.chrXL_group1e 8189033 110 + 12523060 --GCCCCACAAUGUACUUCCACGGCCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAGCUUAAUUAAAAUGUGUGCAUUAAA --.......(((((((......(((.....))).......----......(((((((.((((((..(.(((((...--..))))))..))))).).))))))).....)))))))... ( -23.80, z-score = -1.24, R) >droPer1.super_13 1915129 111 - 2293547 -GGCCCCACAAUGUACUUCGACGGCCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAGCUUAAUUAAAAUAUGUGCAUUAAA -........(((((((...((((((.....)))....)))----......(((((((.((((((..(.(((((...--..))))))..))))).).))))))).....)))))))... ( -24.80, z-score = -0.92, R) >droVir3.scaffold_12726 2807643 91 + 2840439 -------------------------UCAUUCUCCAUUGUGUUCUCUCUGAUUAAUUACGUGACGCGAAUGCAAAUA--AAUUGCGGCCCGUUAACUUAAUUAAAAUUUGUGCAUUAAA -------------------------............((((...(.....(((((((..(((((.(..(((((...--..)))))..))))))...))))))).....).)))).... ( -13.40, z-score = -0.30, R) >droMoj3.scaffold_6473 11262860 92 - 16943266 ----------------------UCGCCACUCUCCAUUGUC--CUGUCUGAUUAAUUACGUGACGUGAAUGCAAAUA--AAUUCCGGCCCGUUAACUUAAUUAAAAUUUGUGCAUUAAA ----------------------..((((.....((.....--.))..(((((((..(((.(.((.((((.......--.)))))).).)))....))))))).....)).))...... ( -9.50, z-score = 0.82, R) >droGri2.scaffold_15203 6405668 103 + 11997470 -----------ACCUUUUCCAUUCUGCGCUCUCCAUUGUG--CUCUUUGAUUAAUUACGUGACGAGCAUGCAAAUA--AAUUGCGGAACGUUAACUUAAUUAAAAUUUGUGCAUUAAA -----------........((....((((........)))--)....)).(((((((..(((((..(.(((((...--..))))))..)))))...)))))))............... ( -17.60, z-score = -0.61, R) >consensus __________________CCACAGCC_CCUGCCCAUUGUC____CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA__AAUUGCGGUCCGUUAACUUAAUUAAAAUUUGUGCAUUAAA ..................................................(((((((.((((((....(((((.......)))))...))))).).)))))))............... ( -9.53 = -9.80 + 0.27)



| Location | 8,912,072 – 8,912,169 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Shannon entropy | 0.48091 |
| G+C content | 0.39574 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -9.53 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8912072 97 + 22422827 -----------CCCCUGCCCUUUUUU-----CUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAAACCGAACCCUU- -----------....(((.(.....(-----(((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))................- ( -20.10, z-score = -3.13, R) >droSim1.chrX_random 2618598 98 + 5698898 -----------CCCCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAAACCGAACCCUU- -----------....(((.(......----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))................- ( -19.50, z-score = -2.82, R) >droSec1.super_34 274785 98 + 457129 -----------CCCCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAAACCGAACCCUU- -----------....(((.(......----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))................- ( -19.50, z-score = -2.82, R) >droYak2.chrX 9370033 98 - 21770863 -----------CCCCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAAACUGAACCCUU- -----------....(((.(......----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))................- ( -19.50, z-score = -2.90, R) >droEre2.scaffold_4690 12547194 98 - 18748788 -----------CCCCUGCCCUUUUUU----UCUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAACUUAAUUAAGAUUUGUGCAUUAAAACCGAACCCUU- -----------....(((.(......----((((((((((.....(((((..(.(((((...--..))))))..)))))..))))))))))...).)))................- ( -19.50, z-score = -2.82, R) >droAna3.scaffold_13047 732385 93 + 1816235 ------------UCCUUCCCACUGUC----CUCUGAUUAAUUACGUGACGGGCAUGCAAAUA--AGAGGUGGGCCGGGGACUUAAUUAAGUCCUCUGCUCUGCCGGAUCCU----- ------------(((.......((((----(((.............)).)))))........--...((..(((.((((((((....)))))))).)).)..)))))....----- ( -29.72, z-score = -1.23, R) >droWil1.scaffold_181150 4196261 108 - 4952429 CACUGGUUUCUCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGCGAAUGCAAAUAUAAAUUGCGGCCCGUUAACUUAAUUAAAAUUUGUGCGAAAAACCAAACUC---- ...((((((.((....((.((.....----.(((((((((.....(((((.(..(((((.......)))))..))))))..)))))))))...)).)))).)))))).....---- ( -19.60, z-score = -1.46, R) >dp4.chrXL_group1e 8189050 104 + 12523060 -----CACGGCCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAGCUUAAUUAAAAUGUGUGCAUUAAAGCCGUAUGCCU- -----.(((((....(((.(((....----.(((((((((..(((.(((.((((........--...))))))))))....)))))))))..))).))).....)))))......- ( -25.30, z-score = -1.40, R) >droPer1.super_13 1915147 104 - 2293547 -----GACGGCCUCCUGCCCAUUGUC----CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA--AAUUGCGGUCCGUUAGCUUAAUUAAAAUAUGUGCAUUAAAACCGAAUGCCU- -----((((((.....)))....)))----......(((((((.((((((..(.(((((...--..))))))..))))).).))))))).......(((((.......)))))..- ( -20.60, z-score = -0.64, R) >droVir3.scaffold_12726 2807644 101 + 2840439 ------------CAUUCUCCAUUGUGUUCUCUCUGAUUAAUUACGUGACGCGAAUGCAAAUA--AAUUGCGGCCCGUUAACUUAAUUAAAAUUUGUGCAUUAAAACGUCCAAACC- ------------........................(((((((..(((((.(..(((((...--..)))))..))))))...)))))))..((((..(........)..))))..- ( -14.70, z-score = -0.34, R) >droMoj3.scaffold_6473 11262864 96 - 16943266 ------------CACUCUCCAUUGUC--CUGUCUGAUUAAUUACGUGACGUGAAUGCAAAUA--AAUUCCGGCCCGUUAACUUAAUUAAAAUUUGUGCAUUAAAAUAGCACC---- ------------..............--.....(((((((..(((.(.((.((((.......--.)))))).).)))....)))))))......((((.........)))).---- ( -12.60, z-score = -0.12, R) >droGri2.scaffold_15203 6405674 109 + 11997470 ---UCCAUUCUGCGCUCUCCAUUGUG--CUCUUUGAUUAAUUACGUGACGAGCAUGCAAAUA--AAUUGCGGAACGUUAACUUAAUUAAAAUUUGUGCAUUAAAAUAGCGCCCCAC ---........(((((.......(((--(.(.....(((((((..(((((..(.(((((...--..))))))..)))))...))))))).....).))))......)))))..... ( -24.22, z-score = -1.99, R) >consensus ___________CCCCUGCCCAUUGUC____CUUUGAUUAAUUACGUGACGUGCAUGCAAAUA__AAUUGCGGUCCGUUAACUUAAUUAAAAUUUGUGCAUUAAAACCGAACCCUU_ ....................................(((((((.((((((....(((((.......)))))...))))).).)))))))........................... ( -9.53 = -9.80 + 0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:10 2011