| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,887,353 – 8,887,451 |
| Length | 98 |
| Max. P | 0.538010 |

| Location | 8,887,353 – 8,887,451 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.75 |
| Shannon entropy | 0.48371 |
| G+C content | 0.41904 |
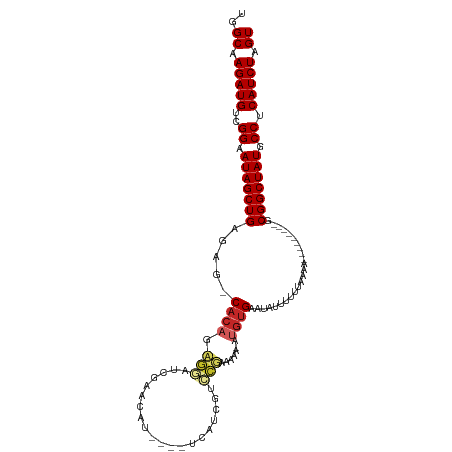
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8887353 98 - 22422827 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGCGGAUCGAACAU---UUUAUCGUCCGAAAAA-AUGUGAUUAUUUUUGAAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-.....((((.(((....---....))))))).((((-(((....))))))).....---------.))))))).)).))))).)) ( -26.60, z-score = -1.46, R) >droSim1.chrX 7083983 98 - 17042790 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGCGGAUCGAGCCU---CUUAUCGUCCGAAAAA-AUGUGAUUAUUUUUGAAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-.....((((.(((....---....))))))).((((-(((....))))))).....---------.))))))).)).))))).)) ( -27.40, z-score = -1.01, R) >droSec1.super_34 250199 98 - 457129 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGCGGAUCGAGCCU---CUUAUCGUCCGAAAAA-AUGUGAUUAUUUUUGAAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-.....((((.(((....---....))))))).((((-(((....))))))).....---------.))))))).)).))))).)) ( -27.40, z-score = -1.01, R) >droYak2.chrX 9345058 98 + 21770863 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGCGGAUCGAGCCU---UUGAUCGUCCGAAAAA-AUGUGAAUAUUUUUUAAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-.....((((.(((....---....)))))))(((((-(((....))))))))....---------.))))))).)).))))).)) ( -27.60, z-score = -1.33, R) >droEre2.scaffold_4690 12523368 98 + 18748788 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGCGGAUCGAGCCU---CUUAUCGUCCGAAAAA-AUGUGAAUAUUUUUGAAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-((((.((((.(((....---....))))))).....-.)))).......((....)---------)))))))).)).))))).)) ( -27.00, z-score = -1.00, R) >droAna3.scaffold_13047 712450 88 - 1816235 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACAGAGGAUCGAGCGA---GUCAUCGACCGAGA------CGAA-----UUCCAAA---------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-......(..(((..(((---....)))..)))..------)...-----.......---------.))))))).)).))))).)) ( -25.70, z-score = -0.63, R) >droWil1.scaffold_181150 4157820 111 + 4952429 UGGCAAGAUGUCGGAAUAGCUGAAAGUCACACAAAAUCAUAAAAUGUUUGCCGUUUUGUUAUUGUUGUGAA-UUUUAUUGAUUUCAAAAAUUACGGCUAUGCCUCAUCUAGU ..((.(((((..((.((((((((((((((((...(((.((((((((.....)))))))).)))..)))).)-)))).(((....)))......))))))).)).))))).)) ( -28.40, z-score = -2.61, R) >droMoj3.scaffold_6473 11232005 98 + 16943266 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACA-AAUAUCAUAGAA----UCCAGAAUUUAUAUU-UUGUGUA-UUUUAUUGAGUACA------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-....-............----..((((((....)))-)))((((-(((....)))))))------.))))))).)).))))).)) ( -24.30, z-score = -1.43, R) >droVir3.scaffold_12726 2775769 97 - 2840439 UGGCAAGAUGUCGGAAUAGCUGAGAG-CACA-AA-GUCAUA-AA----UCCAGAAUUUCUAUU-UUGUGUAUUUUUAUUGAGUACA------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((....-....-..-......-..----..((((((....)))-)))(((((((.....)))))))------.))))))).)).))))).)) ( -24.30, z-score = -1.36, R) >droGri2.scaffold_15203 6378309 97 - 11997470 UGGCAAGAUGUCGGAAUAGCUGACAG-CAC--AAGAUCACAAA-----UCAAGAAACUUUAUU-UUGUGUAUUUUUAUUGAGUACA------GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((.(..-.((--(((((......-----............)))-))))((((((.....)))))).------)))))))).)).))))).)) ( -23.07, z-score = -1.17, R) >consensus UGGCAAGAUGUCGGAAUAGCUGAGAG_CACAGAGGAUCGAACAU____UCAUCGUCCGAAAAA_AUGUGAAUAUUUUUUAAAA_________GCGGCUAUGCCUCAUCUAGU ..((.(((((..((.(((((((.......................................................................))))))).)).))))).)) (-13.57 = -13.57 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:06 2011