
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,880,114 – 8,880,231 |
| Length | 117 |
| Max. P | 0.864937 |

| Location | 8,880,114 – 8,880,231 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Shannon entropy | 0.47887 |
| G+C content | 0.42146 |
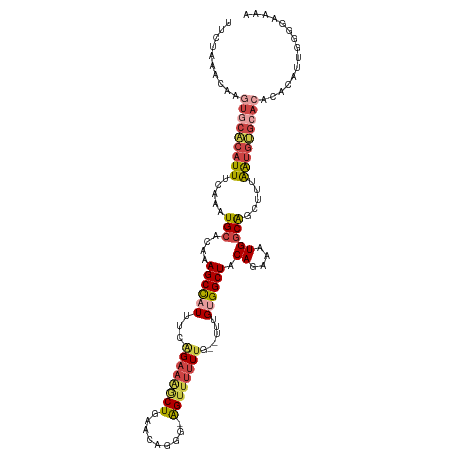
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -17.76 |
| Energy contribution | -20.57 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
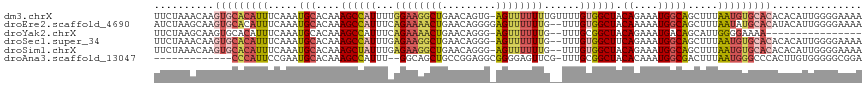
>dm3.chrX 8880114 117 + 22422827 UUCUAAACAAGUGCACAUUUCAAAUGCACAAAGCCAUUUUGGAAGGCUGAACAGUG-AGUUUUUUUGUUUUGUGGCUACAGAAAUGGCAGCUUUAAUGUGCACACACAUUGGGGAAAA ((((...((((((((((((.....(((....((((((...((((((((........-))))))))......)))))).((....))))).....))))))))).....))).)))).. ( -33.40, z-score = -1.73, R) >droEre2.scaffold_4690 12517163 116 - 18748788 AUCUAAGCAAGUGCACAUUUCAAAUGCACAAAGCCAUUUCAGAAAACUGAACAGGGGAGUUUUUUG--UUUGUGGCUACAAAAAUGGCAGCUUUAAUAUGCACAUACAUUGGGGAAAA .(((((....(((((.(((.....(((....((((((..(((((((((.........)))))))))--...)))))).((....))))).....))).))))).....)))))..... ( -29.30, z-score = -1.73, R) >droYak2.chrX 9338225 99 - 21770863 UUCUAAGCAAGUGCACAUUUCAAAUGCACAAAGCCAUUUCAGAAAACUGAACAGGG-AGUUUUUUG--UUUGCGGCUACAGAAAUGACAGCAUUGGGGAAAA---------------- ((((((((..(((((.........)))))..((((....(((((((((........-)))))))))--.....)))).((....))...)).))))))....---------------- ( -24.70, z-score = -1.43, R) >droSec1.super_34 243644 115 + 457129 UUCUAAACAAGUGCACAUUUCAAAUGCACAAAGCCAUUUGAGAAGGCUGAACAGGG-AGUUUUUUG--UUUGUGGCUUCAGAAAUGGCAGCUUUAAUGUGCACACACAUUGGGGAAAA ..........(((((.........)))))...(((((((..((((.((((((((((-....)))))--)))).).))))..)))))))..((((((((((....)))))))))).... ( -35.20, z-score = -2.38, R) >droSim1.chrX 7077839 115 + 17042790 UUCUAAACAAGUGCACAUUUCAAAUGCACAAAGCUAUUUGAGAAGGCUGAACAGGG-AGUUUUUUG--UUUGUGGCUACAGAAAUGGCAGCUUUAAUGUGCACACACAUUGGGGAAAA ((((...((((((((((((((((((((.....)).)))))).(((((((.(((((.-((....)).--))))).....((....)).)))))))))))))))).....))).)))).. ( -32.10, z-score = -2.07, R) >droAna3.scaffold_13047 707298 102 + 1816235 -------------CCCAUUCCGAAUGCACAAAGCCAUUU--GGCAGCUGCCGGAGGCGGGGAGUUCG-UUUGCGGCUACACAAAUGGCGACUUUAAUGGGCCCACUUGUGGGGGCGGA -------------.....((((..(.(((((.(((((((--(......((((.((((((.....)))-))).))))....))))))))........((....)).))))).)..)))) ( -38.10, z-score = -0.48, R) >consensus UUCUAAACAAGUGCACAUUUCAAAUGCACAAAGCCAUUUCAGAAAGCUGAACAGGG_AGUUUUUUG__UUUGUGGCUACAGAAAUGGCAGCUUUAAUGUGCACACACAUUGGGGAAAA ..........(((((((((.....(((....((((((...((((((((.........))))))))......)))))).((....))))).....)))))))))............... (-17.76 = -20.57 + 2.81)
| Location | 8,880,114 – 8,880,231 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Shannon entropy | 0.47887 |
| G+C content | 0.42146 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
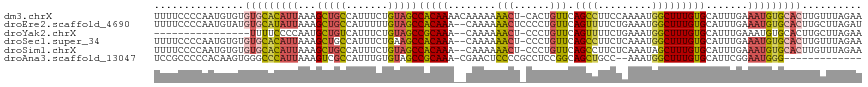
>dm3.chrX 8880114 117 - 22422827 UUUUCCCCAAUGUGUGUGCACAUUAAAGCUGCCAUUUCUGUAGCCACAAAACAAAAAAACU-CACUGUUCAGCCUUCCAAAAUGGCUUUGUGCAUUUGAAAUGUGCACUUGUUUAGAA ...............(((((((((...(((((.......)))))((((((.......(((.-....)))..(((.........))))))))).......))))))))).......... ( -25.90, z-score = -1.06, R) >droEre2.scaffold_4690 12517163 116 + 18748788 UUUUCCCCAAUGUAUGUGCAUAUUAAAGCUGCCAUUUUUGUAGCCACAAA--CAAAAAACUCCCCUGUUCAGUUUUCUGAAAUGGCUUUGUGCAUUUGAAAUGUGCACUUGCUUAGAU ...........(((.((((((((((((((.(((((((((((........)--)))))).........(((((....))))).)))).....)).)))))..))))))).)))...... ( -27.10, z-score = -2.20, R) >droYak2.chrX 9338225 99 + 21770863 ----------------UUUUCCCCAAUGCUGUCAUUUCUGUAGCCGCAAA--CAAAAAACU-CCCUGUUCAGUUUUCUGAAAUGGCUUUGUGCAUUUGAAAUGUGCACUUGCUUAGAA ----------------...........((((.((....)))))).((((.--.........-.((..(((((....)))))..))....((((((.......))))))))))...... ( -19.40, z-score = -0.84, R) >droSec1.super_34 243644 115 - 457129 UUUUCCCCAAUGUGUGUGCACAUUAAAGCUGCCAUUUCUGAAGCCACAAA--CAAAAAACU-CCCUGUUCAGCCUUCUCAAAUGGCUUUGUGCAUUUGAAAUGUGCACUUGUUUAGAA ........((((((....)))))).((((.(((((((..((((.(...((--((.......-...))))..).))))..)))))))...((((((.......))))))..)))).... ( -26.80, z-score = -1.66, R) >droSim1.chrX 7077839 115 - 17042790 UUUUCCCCAAUGUGUGUGCACAUUAAAGCUGCCAUUUCUGUAGCCACAAA--CAAAAAACU-CCCUGUUCAGCCUUCUCAAAUAGCUUUGUGCAUUUGAAAUGUGCACUUGUUUAGAA ...............(((((((((...(((((.......)))))......--.........-...............((((((.((.....))))))))))))))))).......... ( -24.00, z-score = -1.51, R) >droAna3.scaffold_13047 707298 102 - 1816235 UCCGCCCCCACAAGUGGGCCCAUUAAAGUCGCCAUUUGUGUAGCCGCAAA-CGAACUCCCCGCCUCCGGCAGCUGCC--AAAUGGCUUUGUGCAUUCGGAAUGGG------------- ((((....((((((..(((........)))((((((((.(((((((....-))........(((...))).))))))--)))))))))))))....)))).....------------- ( -33.20, z-score = -0.78, R) >consensus UUUUCCCCAAUGUGUGUGCACAUUAAAGCUGCCAUUUCUGUAGCCACAAA__CAAAAAACU_CCCUGUUCAGCCUUCUCAAAUGGCUUUGUGCAUUUGAAAUGUGCACUUGUUUAGAA ...............(((((((((...(((((.......)))))(((((........(((......))).((((.........))))))))).......))))))))).......... (-15.74 = -15.97 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:03 2011