| Sequence ID | dm3.chrX |
|---|---|
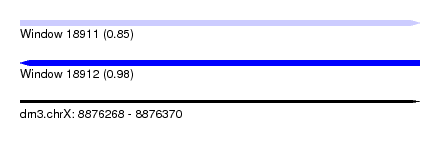
| Location | 8,876,268 – 8,876,370 |
| Length | 102 |
| Max. P | 0.978169 |

| Location | 8,876,268 – 8,876,370 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.42310 |
| G+C content | 0.42547 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852620 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 8876268 102 + 22422827 ------------UAUAG-GUU---AGGUCGCCA-UGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGCCAAUUGAUGG- ------------....(-((.---.....))).-((((..((((((((((((((....(((-((.........))))).-.....))))))))..))))))........))))........- ( -29.90, z-score = -0.89, R) >droSim1.chrX 7074146 102 + 17042790 ------------UAUAG-GUU---AGGUCGCCA-UGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGCCAAUUGAUGG- ------------....(-((.---.....))).-((((..((((((((((((((....(((-((.........))))).-.....))))))))..))))))........))))........- ( -29.90, z-score = -0.89, R) >droSec1.super_34 240052 102 + 457129 ------------UAUAG-GUA---AGGUCGCCA-UGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGCCAAUUGAUGG- ------------....(-(((---.....((..-..))..((((((((((((((....(((-((.........))))).-.....))))))))..)))))).......)))).........- ( -30.10, z-score = -0.94, R) >droYak2.chrX 9334550 102 - 21770863 ------------UAUAG-GUU---AGGUCGCCA-UGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGCUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAAGCCAAUUGUCGG- ------------....(-((.---.....))).-((((((((((((((((((((....(((-((.........))))).-.....))))))))..))))))......))))))........- ( -32.30, z-score = -1.62, R) >droEre2.scaffold_4690 12513571 102 - 18748788 ------------UAUAG-GUU---AGGCCGCUA-UGGCGUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGCCAAUUGUUGG- ------------....(-(..---...))....-((((((((((((((((((((....(((-((.........))))).-.....))))))))..))))))......))))))........- ( -31.50, z-score = -0.92, R) >droAna3.scaffold_13047 704293 100 + 1816235 ------------UAUAG-GUU---GGGUCGCC---GGCUCUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGUCGCAUAUUUUAUGCCGAUUGCUGC- ------------....(-(((---((....))---))))..(((((((((((((....(((-((.........))))).-.....))))))))))((((((((...)))).)))))))...- ( -30.20, z-score = -0.84, R) >droPer1.super_13 1878191 106 - 2293547 ------------UGUGG-GUUUUGGG-UCGCCUCUGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUGUGGCAUGCAAGCAUCGCAUAUUUUAUGCCCAAUUGCUGG ------------....(-((.(((((-(.(((...)))..((((((((((((((....(((-((.........))))).-.....))))))))))..))))........))))))..))).. ( -34.80, z-score = -1.77, R) >dp4.chrXL_group1e 8152027 99 + 12523060 ------------UGAGG-GUUUUGGGGUCGCCUCUGGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAAUAAGU-CUGUGGCAUGCAAGCAUCGCAUAUUUUAUGCCCA-------- ------------...((-((..((((((.(((...)))..((((((((((((((....(((-((.........))))).-.....))))))))))..)))).)))))).)))).-------- ( -32.40, z-score = -1.59, R) >droWil1.scaffold_181150 4141754 102 - 4952429 ------------UAUAG-GUUGUUGU-GCGCCUUUAGGUUUGCGGCUUGCAUGCAAAUUUUUAUGUUUUAAUAAUAAGU-CUGAGGCAUGCAAGCAGCGUAUAUUUUAUGCCAAUUG----- ------------....(-((...(((-(((((....)))..((.((((((((((.....(((((.........))))).-.....)))))))))).)))))))......))).....----- ( -31.10, z-score = -1.98, R) >droVir3.scaffold_12726 2761225 116 + 2840439 AAUGGGCGUGGCUUUGGCGUCUGCUUGAGUCGCUUAGCUUUGCGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAACAAGUUCUCUGGCAUGCAAGCGGCGCAUAUUUUAUGCCAAUUG----- ....(((((((.....(((.(((((((((((((........)))))))((((((.......-.((((.....)))).........))))))))))))))).....))))))).....----- ( -42.83, z-score = -2.91, R) >droMoj3.scaffold_6473 11220908 115 - 16943266 AAUGGGCGUGGUUUUGGCGUCUGCUUGAGUCGCUUAGCUUUGUGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAACAAGU-CUCUGGCAUGCAAGCGGCGCAUAUUUUAUGCCAAUUG----- ....(((((((.....(((.(((((((((((((........)))))))((((((.......-.((((.....))))...-.....))))))))))))))).....))))))).....----- ( -42.19, z-score = -3.26, R) >droGri2.scaffold_15203 6366978 115 + 11997470 AAUGGGAAUGGCUCUGGCGUCUGCUUGAGUCUUUUAGCCCUGUGGCUUGCAUGCAAAUUUU-AUGUUUUAAUAACAAGU-CUCUGGCAUGCAAGCGGCCCAUAUUUUAUGCCAAUGG----- ...(((...(((((.(((....))).)))))......))).((.((((((((((.......-.((((.....))))...-.....)))))))))).))((((...........))))----- ( -37.49, z-score = -1.76, R) >consensus ____________UAUAG_GUU___AGGUCGCCA_UGGCUUUGCGGCUUGCAUGCAAAUUUU_AUGUUUUAAUAAUAAGU_CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGCCAAUUG__GG_ ........................................((((((((((((((.........((((.....)))).........))))))))))..))))..................... (-21.39 = -21.23 + -0.17)

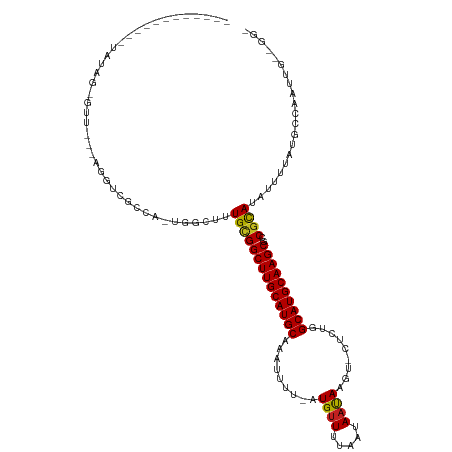
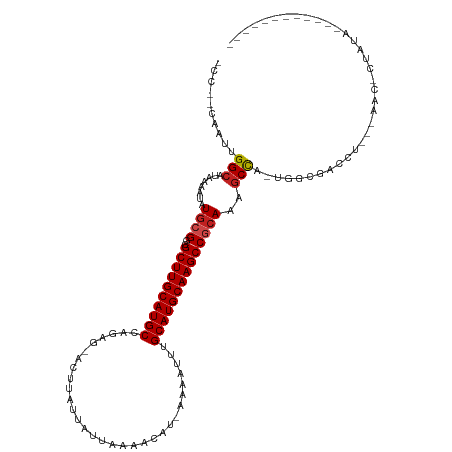
| Location | 8,876,268 – 8,876,370 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.42310 |
| G+C content | 0.42547 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978169 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 8876268 102 - 22422827 -CCAUCAAUUGGCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCCA-UGGCGACCU---AAC-CUAUA------------ -.........(((........((((((...(((((((.(((.-..((((.........))-))..)))))))))).))))))..((..-..))).)).---...-.....------------ ( -25.80, z-score = -1.41, R) >droSim1.chrX 7074146 102 - 17042790 -CCAUCAAUUGGCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCCA-UGGCGACCU---AAC-CUAUA------------ -.........(((........((((((...(((((((.(((.-..((((.........))-))..)))))))))).))))))..((..-..))).)).---...-.....------------ ( -25.80, z-score = -1.41, R) >droSec1.super_34 240052 102 - 457129 -CCAUCAAUUGGCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCCA-UGGCGACCU---UAC-CUAUA------------ -.........(((........((((((...(((((((.(((.-..((((.........))-))..)))))))))).))))))..((..-..))).)).---...-.....------------ ( -25.80, z-score = -1.35, R) >droYak2.chrX 9334550 102 + 21770863 -CCGACAAUUGGCUUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAGCAU-AAAAUUUGCAUGCAAGCCGCAAAGCCA-UGGCGACCU---AAC-CUAUA------------ -.((.(...((((((......((((((...(((((((.(((.-..((((.........))-))..)))))))))).))))))))))))-..)))....---...-.....------------ ( -29.70, z-score = -2.24, R) >droEre2.scaffold_4690 12513571 102 + 18748788 -CCAACAAUUGGCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAACGCCA-UAGCGGCCU---AAC-CUAUA------------ -.........(((........((((((...(((((((.(((.-..((((.........))-))..)))))))))).)))))).(((..-..)))))).---...-.....------------ ( -29.70, z-score = -2.56, R) >droAna3.scaffold_13047 704293 100 - 1816235 -GCAGCAAUCGGCAUAAAAUAUGCGACGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAGAGCC---GGCGACCC---AAC-CUAUA------------ -...((...((((........((((..((((((((((.(((.-..((((.........))-))..)))))))))))))))))..)))---))).....---...-.....------------ ( -26.40, z-score = -1.99, R) >droPer1.super_13 1878191 106 + 2293547 CCAGCAAUUGGGCAUAAAAUAUGCGAUGCUUGCAUGCCACAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCCAGAGGCGA-CCCAAAAC-CCACA------------ .......((((((........((((..((((((((((.....-.................-.......))))))))))))))..(((...)))).-)))))...-.....------------ ( -29.55, z-score = -2.87, R) >dp4.chrXL_group1e 8152027 99 - 12523060 --------UGGGCAUAAAAUAUGCGAUGCUUGCAUGCCACAG-ACUUAUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCCAGAGGCGACCCCAAAAC-CCUCA------------ --------((((.........((((..((((((((((.....-.................-.......))))))))))))))..(((...)))...))))....-.....------------ ( -27.45, z-score = -2.70, R) >droWil1.scaffold_181150 4141754 102 + 4952429 -----CAAUUGGCAUAAAAUAUACGCUGCUUGCAUGCCUCAG-ACUUAUUAUUAAAACAUAAAAAUUUGCAUGCAAGCCGCAAACCUAAAGGCGC-ACAACAAC-CUAUA------------ -----...(((((...........((.((((((((((.....-.........................)))))))))).))...((....)).))-.)))....-.....------------ ( -22.61, z-score = -2.13, R) >droVir3.scaffold_12726 2761225 116 - 2840439 -----CAAUUGGCAUAAAAUAUGCGCCGCUUGCAUGCCAGAGAACUUGUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCGCAAAGCUAAGCGACUCAAGCAGACGCCAAAGCCACGCCCAUU -----.....(((........((((..((((((((((.(((.....((((.....)))).-....)))))))))))))))))..(((..(((.((.....)).)))...)))...))).... ( -34.50, z-score = -3.04, R) >droMoj3.scaffold_6473 11220908 115 + 16943266 -----CAAUUGGCAUAAAAUAUGCGCCGCUUGCAUGCCAGAG-ACUUGUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCACAAAGCUAAGCGACUCAAGCAGACGCCAAAACCACGCCCAUU -----...(((((.........((...((((((((((.(((.-...((((.....)))).-....)))))))))))))......))...((.......))....)))))............. ( -29.50, z-score = -2.40, R) >droGri2.scaffold_15203 6366978 115 - 11997470 -----CCAUUGGCAUAAAAUAUGGGCCGCUUGCAUGCCAGAG-ACUUGUUAUUAAAACAU-AAAAUUUGCAUGCAAGCCACAGGGCUAAAAGACUCAAGCAGACGCCAGAGCCAUUCCCAUU -----....((((........(((.((((((((((((.(((.-...((((.....)))).-....)))))))))))))....)).)))......((..((....))..))))))........ ( -31.20, z-score = -1.54, R) >consensus _CC__CAAUUGGCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG_ACUUAUUAUUAAAACAU_AAAAUUUGCAUGCAAGCCGCAAAGCCA_UGGCGACCU___AAC_CUAUA____________ ..........(((........((((..((((((((((...............................))))))))))))))..)))................................... (-18.41 = -18.52 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:27:01 2011