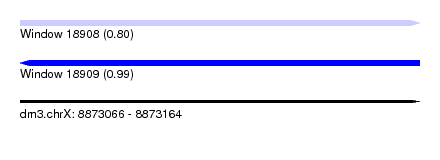
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,873,066 – 8,873,164 |
| Length | 98 |
| Max. P | 0.985318 |

| Location | 8,873,066 – 8,873,164 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.34 |
| Shannon entropy | 0.44930 |
| G+C content | 0.39274 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.53 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
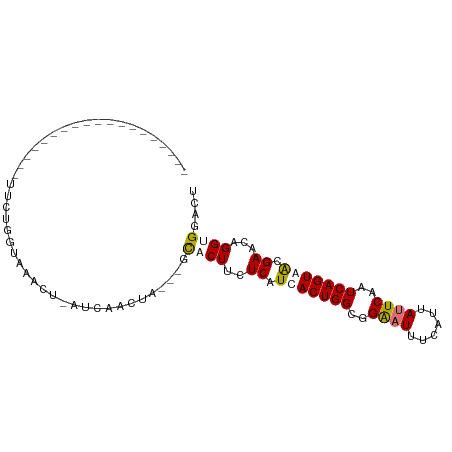
>dm3.chrX 8873066 98 + 22422827 UUUUGGGUUAGCUCAUCAACUUUUGGAAACU-AUCAACUAUCAGCUCUACUCAUCACUGGCGGAAUUUCAUUAUUCAAUCAGUAGCGAACAGGUGGACU ...(((((.((((...........(....).-..........))))..)))))..(((((..((((......))))..)))))................ ( -16.85, z-score = -0.40, R) >droEre2.scaffold_4690 12509638 93 - 18748788 -UUCUGGUUAGUUAAUAAAUUUUGGAAAACG-AU-AACUA---GUUCUUCUCAUCACUGGCGGGCUUUAAUUAUUCAAUCAGUAGCGAACAGGUGAACU -..(((((((.....................-.)-)))))---).........(((((..((.(((..............)))..))....)))))... ( -10.59, z-score = 1.65, R) >droSec1.super_34 235449 77 + 457129 -------------------UUCUGGUUAGUUCAUCAACCA---GCACUUCUCAUCACUGGCGGAAUUUCAUUAUUCAAUCAGUGACGAACAGGUGGAAA -------------------..((((((........)))))---)(((((.((.(((((((..((((......))))..))))))).))..))))).... ( -23.30, z-score = -3.64, R) >droSim1.chrX 7071398 77 + 17042790 -------------------UUCUGGUUAGUUCAUCAACUA---GCACUUCUCAUCACUGGCGGAAUUUUAUUAUUCAAUCAGUGACGAACAGGUGGACU -------------------.(((((((((((....)))))---)).....((.(((((((..((((......))))..))))))).)).))))...... ( -22.10, z-score = -3.57, R) >consensus ___________________UUCUGGUAAACU_AUCAACUA___GCACUUCUCAUCACUGGCGGAAUUUCAUUAUUCAAUCAGUAACGAACAGGUGGACU ............................................((((..((.(((((((..((((......))))..))))))).))...)))).... (-10.90 = -11.53 + 0.63)

| Location | 8,873,066 – 8,873,164 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.34 |
| Shannon entropy | 0.44930 |
| G+C content | 0.39274 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -11.45 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
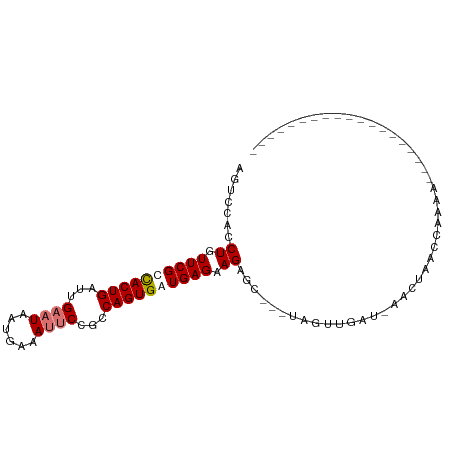
>dm3.chrX 8873066 98 - 22422827 AGUCCACCUGUUCGCUACUGAUUGAAUAAUGAAAUUCCGCCAGUGAUGAGUAGAGCUGAUAGUUGAU-AGUUUCCAAAAGUUGAUGAGCUAACCCAAAA .......(((((((.(((((...((((......))))...))))).)))))))((((.((..(((..-......)))......)).))))......... ( -15.90, z-score = -0.64, R) >droEre2.scaffold_4690 12509638 93 + 18748788 AGUUCACCUGUUCGCUACUGAUUGAAUAAUUAAAGCCCGCCAGUGAUGAGAAGAAC---UAGUU-AU-CGUUUUCCAAAAUUUAUUAACUAACCAGAA- .((((.....((((.(((((....................))))).))))..))))---(((((-(.-.((((....))))....)))))).......- ( -10.35, z-score = 0.44, R) >droSec1.super_34 235449 77 - 457129 UUUCCACCUGUUCGUCACUGAUUGAAUAAUGAAAUUCCGCCAGUGAUGAGAAGUGC---UGGUUGAUGAACUAACCAGAA------------------- ....(((...((((((((((...((((......))))...))))))))))..)))(---((((((......)))))))..------------------- ( -25.30, z-score = -4.92, R) >droSim1.chrX 7071398 77 - 17042790 AGUCCACCUGUUCGUCACUGAUUGAAUAAUAAAAUUCCGCCAGUGAUGAGAAGUGC---UAGUUGAUGAACUAACCAGAA------------------- ....(((...((((((((((...((((......))))...))))))))))..))).---(((((....))))).......------------------- ( -19.30, z-score = -3.38, R) >consensus AGUCCACCUGUUCGCCACUGAUUGAAUAAUGAAAUUCCGCCAGUGAUGAGAAGAGC___UAGUUGAU_AACUAACCAAAA___________________ .......((.((((((((((...((((......))))...)))))))))).)).............................................. (-11.45 = -12.20 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:59 2011