
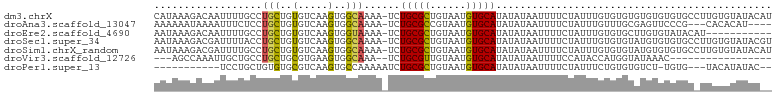
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,836,839 – 8,836,941 |
| Length | 102 |
| Max. P | 0.611728 |

| Location | 8,836,839 – 8,836,941 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.29 |
| Shannon entropy | 0.54876 |
| G+C content | 0.36961 |
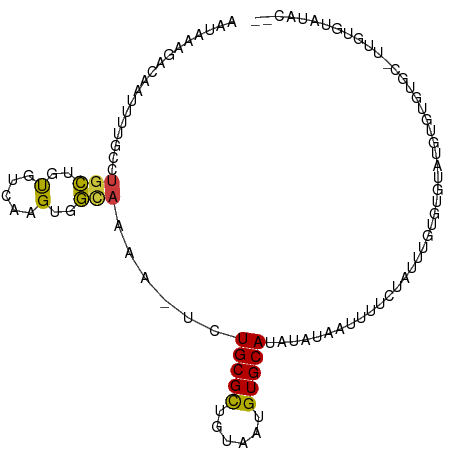
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.16 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8836839 102 + 22422827 CAUAAAGACAAUUUUGCCUGCUGUGUCAAGUGGCAAAA-UCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUGUGUGUGUGUGUGUGCCUUGUGUAUACAU ..........((((((((.(((......))))))))))-).(((((.((((((..(((((((((........)))))))))..))).)))...)))))..... ( -27.20, z-score = -1.84, R) >droAna3.scaffold_13047 675753 95 + 1816235 AAAAAAUAAAAUUUCUCCUGCUGUGUCAAGUGGCAAAA-UCUGCGCCGUAAUGUGCAUAUAUAAUUUUCUAUUUGUUUGCGAGUUCCCG---CACACAU---- .....................((((((((((((.((((-(.(((((......)))))......))))))))))))...(((......))---)))))).---- ( -17.60, z-score = -0.98, R) >droEre2.scaffold_4690 12481264 91 - 18748788 AAUAAAGACAAUUUUGCCUGCUGUGUCAAGUGGUAAAA-UCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUGUGUGCUUGUGUAUACAU----------- .....(((..((((((((.(((......))))))))))-).(((((......)))))..........)))...(((((((....))))))).----------- ( -22.40, z-score = -2.12, R) >droSec1.super_34 207742 102 + 457129 AAUAAAGACGAUUUUACCUGCUGUGUCAAGUGGCAAAA-UCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUGUGUGUAUGUGUGUGCCUUGUGUAUACGU .........((((((.((.(((......))))).))))-)).((((......))))((((((((........))))))))(((((((..(...)..))))))) ( -23.70, z-score = -1.19, R) >droSim1.chrX_random 2613932 102 + 5698898 AAUAAAGACGAUUUUGCCUGCUGUGUCAAGUGGCAAAA-UCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUGUGUGUAUGUGUGUGCCUUGUGUAUACAU .........(((((((((.(((......))))))))))-)).((((......))))((((((((........))))))))..((((((((.....)))))))) ( -30.90, z-score = -3.65, R) >droVir3.scaffold_12726 2710400 81 + 2840439 ---AGCCAAAUUGCUGCCUGCUGCGUGAAGUGGCAAA--UCUGCGUUGUAAUGUGCAUAUAUAAUUUUCCAUACCAUGGUAUAAAC----------------- ---.((...((((((((.(((..(.....)..)))..--...)))..)))))..))..............((((....))))....----------------- ( -14.60, z-score = 1.21, R) >droPer1.super_13 1842692 86 - 2293547 -----------UCCUGCUGUGUGCGUCAAGUGCCAAAAAUCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUCUGUGUGUCU-UGUG---UACAUAUAC-- -----------......((((((((.((((............((((......))))(((((((............)))))))))-))))---))))))...-- ( -18.20, z-score = -1.71, R) >consensus AAUAAAGACAAUUUUGCCUGCUGUGUCAAGUGGCAAAA_UCUGCGCUGUAAUGUGCAUAUAUAAUUUUCUAUUUGUGUGUAUGUGUGUGC_UUGUGUAUAC__ ..................(((..(.....)..)))......(((((......))))).............................................. ( -9.54 = -9.16 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:56 2011