| Sequence ID | dm3.chr2L |
|---|---|
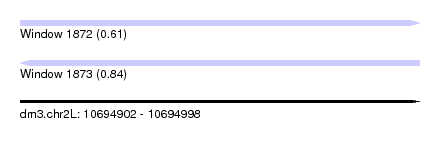
| Location | 10,694,902 – 10,694,998 |
| Length | 96 |
| Max. P | 0.841861 |

| Location | 10,694,902 – 10,694,998 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.44 |
| Shannon entropy | 0.50282 |
| G+C content | 0.35275 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -8.57 |
| Energy contribution | -8.71 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10694902 96 + 23011544 GGUUACGACAAUGACCCUUGGGGUCUACGUCACUC-GGUUUGAUGGAGUGAAAAUACAGACGCCAC-ACAGAGUGAUGAAUUAUAUAAAAUUAUUAAA------------- ............(((((...)))))..((((((((-....((.(((.((..........)).))))-)..))))))))....................------------- ( -22.10, z-score = -1.06, R) >droSim1.chr2L 10493789 97 + 22036055 GGUUACGACAAUGACCUUUGGGGUCUACGUCACUCUGGUUUGAUGGAGUGAAAAUACAGACGCCAC-ACAGAGUGAUGAAUUAUAUAAAAUUAUUAAA------------- ............((((.....))))..((((((((((...((.(((.((..........)).))))-)))))))))))....................------------- ( -23.80, z-score = -1.85, R) >droSec1.super_3 6112579 97 + 7220098 GGUUACGACAAUGACCUUUGGGGUCUACGUCACUCUGGUUUGAUGGAGUGAAAACACAGACGCCAC-ACAGAGUGAUGAAUUAUAUAAAAUUAUUAAA------------- ............((((.....))))..((((((((((...((.(((.(((....))).....))))-)))))))))))....................------------- ( -24.20, z-score = -1.85, R) >droEre2.scaffold_4929 11904058 97 - 26641161 GGUUACGACAAUGACCCUUAGGGUCUACGUCACUCUGGUUUGAUGGGGUGGCAAUAUAGACGCAAC-ACAGAGUGAUGAAUUAUAUAAAAUUAUUAAA------------- ............(((((...)))))..((((((((((..(((.((..(((....)))...))))).-.))))))))))....................------------- ( -25.70, z-score = -2.26, R) >droAna3.scaffold_12916 14538142 96 + 16180835 GUUUGCGCCAUUGACCCA-AAGGUCUACGUCACUCUAUCACAAUAGAUUAA-AAUACAAUCGAAAUUAAAAGGUAAUGAAUUAUAUAAAAUUACUUAA------------- ....(((.....((((..-..))))..)))...(((((....)))))....-..................(((((((............)))))))..------------- ( -10.10, z-score = 0.06, R) >dp4.chr4_group4 2900147 107 + 6586962 GGUUACGACAAUGACCCUAAAGGUCUACGUCAUUCAAUCUGUAUAGAUUAA-AAUACAAUCGUAUU---ACAAUCAUCAGUCAUGUAACAAAAAUACAUAUGCCUAAUAAA .((((((((...((((.....))))...))).......(((.((.((((..-(((((....)))))---..)))))))))....)))))...................... ( -19.90, z-score = -2.79, R) >droPer1.super_10 1914005 107 + 3432795 GGUUACGACAAUGACCCUAAAGGUCUACGUCAUUCAAUCUGUAUAGAUUAA-AAUACAAUCGUAUU---ACAAUCAUCAGUCAUGUAACAAAAAUACAUAUGCCUAAUAAA .((((((((...((((.....))))...))).......(((.((.((((..-(((((....)))))---..)))))))))....)))))...................... ( -19.90, z-score = -2.79, R) >consensus GGUUACGACAAUGACCCUUAGGGUCUACGUCACUCUGGUUUGAUGGAGUGA_AAUACAGACGCAAC_ACAGAGUGAUGAAUUAUAUAAAAUUAUUAAA_____________ ......(((...((((.....))))...)))................................................................................ ( -8.57 = -8.71 + 0.14)



| Location | 10,694,902 – 10,694,998 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.44 |
| Shannon entropy | 0.50282 |
| G+C content | 0.35275 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -9.91 |
| Energy contribution | -9.73 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
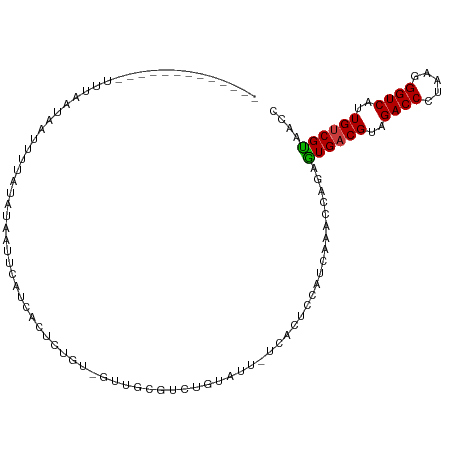
>dm3.chr2L 10694902 96 - 23011544 -------------UUUAAUAAUUUUAUAUAAUUCAUCACUCUGU-GUGGCGUCUGUAUUUUCACUCCAUCAAACC-GAGUGACGUAGACCCCAAGGGUCAUUGUCGUAACC -------------..........(((((((((...((((((.((-((((.((..........)).))))...)).-))))))....(((((...)))))))))).)))).. ( -19.80, z-score = -1.30, R) >droSim1.chr2L 10493789 97 - 22036055 -------------UUUAAUAAUUUUAUAUAAUUCAUCACUCUGU-GUGGCGUCUGUAUUUUCACUCCAUCAAACCAGAGUGACGUAGACCCCAAAGGUCAUUGUCGUAACC -------------..........(((((((((...(((((((((-((((.((..........)).))).))...))))))))....((((.....))))))))).)))).. ( -19.60, z-score = -1.87, R) >droSec1.super_3 6112579 97 - 7220098 -------------UUUAAUAAUUUUAUAUAAUUCAUCACUCUGU-GUGGCGUCUGUGUUUUCACUCCAUCAAACCAGAGUGACGUAGACCCCAAAGGUCAUUGUCGUAACC -------------..........(((((((((...((((((((.-((.......(((....)))........))))))))))....((((.....))))))))).)))).. ( -21.16, z-score = -2.05, R) >droEre2.scaffold_4929 11904058 97 + 26641161 -------------UUUAAUAAUUUUAUAUAAUUCAUCACUCUGU-GUUGCGUCUAUAUUGCCACCCCAUCAAACCAGAGUGACGUAGACCCUAAGGGUCAUUGUCGUAACC -------------..........(((((((((...((((((((.-((((((.......)))..........)))))))))))....(((((...)))))))))).)))).. ( -21.00, z-score = -2.36, R) >droAna3.scaffold_12916 14538142 96 - 16180835 -------------UUAAGUAAUUUUAUAUAAUUCAUUACCUUUUAAUUUCGAUUGUAUU-UUAAUCUAUUGUGAUAGAGUGACGUAGACCUU-UGGGUCAAUGGCGCAAAC -------------............((((((((.((((.....))))...)))))))).-....(((((....)))))(((.(((.((((..-..)))).))).))).... ( -15.30, z-score = -0.20, R) >dp4.chr4_group4 2900147 107 - 6586962 UUUAUUAGGCAUAUGUAUUUUUGUUACAUGACUGAUGAUUGU---AAUACGAUUGUAUU-UUAAUCUAUACAGAUUGAAUGACGUAGACCUUUAGGGUCAUUGUCGUAACC .(((((((...((((((.......)))))).)))))))....---..((((((.((..(-(((((((....))))))))..))...((((.....))))...))))))... ( -26.10, z-score = -2.43, R) >droPer1.super_10 1914005 107 - 3432795 UUUAUUAGGCAUAUGUAUUUUUGUUACAUGACUGAUGAUUGU---AAUACGAUUGUAUU-UUAAUCUAUACAGAUUGAAUGACGUAGACCUUUAGGGUCAUUGUCGUAACC .(((((((...((((((.......)))))).)))))))....---..((((((.((..(-(((((((....))))))))..))...((((.....))))...))))))... ( -26.10, z-score = -2.43, R) >consensus _____________UUUAAUAAUUUUAUAUAAUUCAUCACUCUGU_GUUGCGUCUGUAUU_UCACUCCAUCAAACCAGAGUGACGUAGACCCUAAGGGUCAUUGUCGUAACC ..............................................................................((((((..((((.....))))..)))))).... ( -9.91 = -9.73 + -0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:43 2011