
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,800,972 – 8,801,070 |
| Length | 98 |
| Max. P | 0.515950 |

| Location | 8,800,972 – 8,801,070 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 56.43 |
| Shannon entropy | 0.86603 |
| G+C content | 0.55766 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -5.62 |
| Energy contribution | -6.58 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8800972 98 - 22422827 AUCA--AACAGCUGGUGGGCACUCGGAUCUUCGAUUGCCGCUGUCUGGCAGCACUGGGAACAGUGAGAACAGUGGCCAGACUGCG-GAGC-------UGGCGAUGGGC (((.--..(((((....((((.((((....)))).))))((.(((((((..(((((....))))).........))))))).)).-.)))-------))..))).... ( -41.90, z-score = -2.23, R) >droSim1.chrX 7013455 97 - 17042790 AUCA--AACAGCUGGUGGGCACUCGGAUCGCCGAUUGCCGCUGUCUGGCAGCACUGGG--CCGUGAGCAAAGUGGCCAGACUGCGAGAGC-------UGGCGAUGGGC (((.--..(((((....((((.((((....)))).))))((.(((((((..((((..(--.(....))..))))))))))).))...)))-------))..))).... ( -44.10, z-score = -1.58, R) >droSec1.super_34 171346 97 - 457129 AUCA--AACAGCUGGUGGGCACUCGGAUCGCCGAUUGCCGCUGUCUGGCAGCGCUGGG--CCGUGAGAACAGUGGCCAGACUGCGAGAGC-------UGGCGAUGGGC (((.--..(((((....((((.((((....)))).))))((.(((((((..(((((..--.(....)..)))))))))))).))...)))-------))..))).... ( -45.00, z-score = -1.69, R) >droYak2.chrX 9268525 97 + 21770863 AUCA--AACAGCUGGUGAGCACUCGGAUCGCAGAUUGCCGCUGUCUGGCAACACUGAG--CAGUGGAAAAGAUGGCCAGACUGCGAGAGC-------UUGCGAUGAGC ....--....(((....))).(((..(((((((.((..(((.(((((((..((((...--.)))).........))))))).))).))..-------)))))))))). ( -33.90, z-score = -0.76, R) >droEre2.scaffold_4690 12448072 104 + 18748788 AUCA--AACAGCUGGUGGCCACUCCGAUCGCCGAUUUCCGCUGUCUGGCAACACUGGG--CAGUGGGAAAAGUGGCAAGGCUGCGAGAGCGAUAUUAUUGCGAUGGGC (((.--..(((((....((((((.((.....)).(((((((((((..(.....)..))--))))))))).))))))..))))).....(((((...)))))))).... ( -40.60, z-score = -1.76, R) >droAna3.scaffold_13047 646717 87 - 1816235 AUCAGCAGCAAGAGAUGGCCACUUG--------GCGUCCGCUGAGGUGCAAUAUCA----CCGCCA-UAGAGAUGAGAGAUUGUGAAC--------UUAUCGAUCCUC ...........((((((((...(..--------((....))..)((((......))----))))))-)...((((((..........)--------)))))....))) ( -21.20, z-score = 0.36, R) >dp4.chrXL_group1e 8080339 96 - 12523060 ACAA--UACAACAAAUAAACAGCUGGA------GUUUCGCCGAUCUGGCAGCGCGAGAACCCAUCA-UAGAGCUGCCAGACUAAAGGAAAAA---GAUUCUGCUCAGC ....--..............(((.(((------((((..((..(((((((((.(..((.....)).-..).))))))))).....))....)---))))))))).... ( -25.70, z-score = -2.02, R) >droPer1.super_13 1807623 96 + 2293547 ACAA--UACAACAAAUAAACAGCUGGA------GUUUCGCCGAUCUGGCAGCGCGAGAACCCAUCA-UAGAGCUGCCAGACUAAAGGAAAAA---GAUUCUGCCCAGC ....--...............((((((------(..((.((..(((((((((.(..((.....)).-..).))))))))).....)).....---))..))..))))) ( -27.40, z-score = -2.86, R) >droVir3.scaffold_12726 2654393 89 - 2840439 AUCA--GCUGUCUAACUGGCUGUUUAAUGUCUAGGUGGCAACGCCAAUUA--ACCAGC--CAGUAGCCAGCGCUGCCAGACAGCUGGC---------UAGCGGC---- ....--((((.(((.(((((((.(((((.....((((....)))).))))--).))))--)))))).))))((((((((....)))))---------.)))...---- ( -44.40, z-score = -4.45, R) >consensus AUCA__AACAGCUGGUGGGCACUCGGAUCGCCGAUUGCCGCUGUCUGGCAGCACUGGG__CCGUGAGAAGAGUGGCCAGACUGCGAGAGC_______UUGCGAUGGGC ..........................................(((((((.((...................)).)))))))........................... ( -5.62 = -6.58 + 0.95)
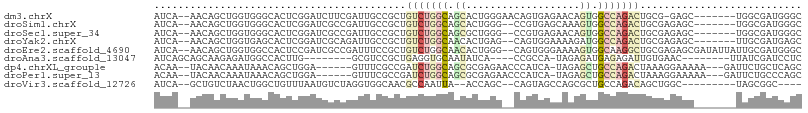

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:53 2011