
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,790,870 – 8,791,026 |
| Length | 156 |
| Max. P | 0.886767 |

| Location | 8,790,870 – 8,790,979 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.40862 |
| G+C content | 0.49627 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.17 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
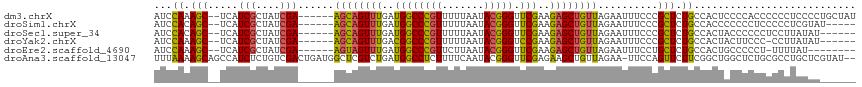
>dm3.chrX 8790870 109 - 22422827 AUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUUUCCCGCUCUGCCACUCCCCACCCCCCUCCCCUGCUAU ......(((--.....))).....------((((((((((..((((((.......)))))))))))(((.((.((((.........)))).)))))..............))))).. ( -24.10, z-score = -1.26, R) >droSim1.chrX 7008064 104 - 17042790 AUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUUUCCCGCUCUGCCACCCCCCCUCCCCCUCGUAU----- ....(((((--((.((((((((((------(....)))))))))((((.......))))....)).))))))).......................................----- ( -26.60, z-score = -2.38, R) >droSec1.super_34 161540 103 - 457129 AUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUUUCCCGCUCUGCCACUACCCCCCUCCUUAUAU------ ....(((((--((.((((((((((------(....)))))))))((((.......))))....)).)))))))......................................------ ( -26.60, z-score = -2.52, R) >droYak2.chrX 9258594 102 + 21770863 AUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGACGGCCCGUUUUUAAUACGGGUCCGAAGAGCUGUUAGAAUUUCCCGCUCUGCCACUACUUCCC-CCUUAUAU------ ...((.(((--...(((....)))------((((((((..((((((((.......))))).)))..))))))))..........))).))............-........------ ( -26.30, z-score = -2.29, R) >droEre2.scaffold_4690 12438699 100 + 18748788 AUCCAAAGC--UCAUCGCUAUCGA------AGUAGUUUGAUGGCCCGUUCUUAAUACGGGUUCGAAGAGCUGUUAGAAUUUCCUGCUCUGCCACUGCCCCCU-UUUUAU-------- ......(((--((.((((((((((------(....)))))))))((((.......))))....)).)))))..(((((......((.........)).....-))))).-------- ( -23.20, z-score = -1.01, R) >droAna3.scaffold_13047 635562 114 - 1816235 UUUAAAAGCAGCCAUCUCUGUCGACUGAUGGCUCGUCUGAUGGCCUCUUUUCAAUACGGGUUCGAGAAGCUGUUAGAA-UUCCAGUUCUUCGGCUGGCUCUGCGCCUGCUCGUAU-- ...(((((..((((((...(.(((........))).).))))))..)))))..((((((((......(((((..((((-(....))))).)))))(((.....))).))))))))-- ( -35.70, z-score = -1.31, R) >consensus AUCCAAAGC__UCAUCGCUAUCGA______AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUUUCCCGCUCUGCCACUACCCCCCCCCCUAUAC______ ...((.(((.....(((....)))......((((((((..((((((((.......))))).)))..))))))))..........))).))........................... (-16.66 = -16.17 + -0.50)
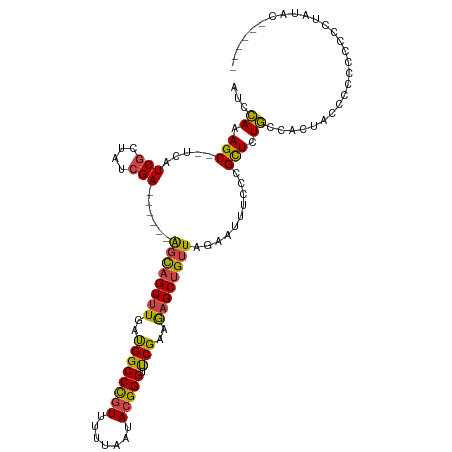

| Location | 8,790,907 – 8,791,019 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Shannon entropy | 0.35134 |
| G+C content | 0.45984 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.07 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.759063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8790907 112 - 22422827 UGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUU ..(((((((.....((.((((....)).)).))....))).)))).(((--((.((((((((((------(....)))))))))((((.......))))....)).)))))......... ( -32.60, z-score = -1.66, R) >droSim1.chrX 7008096 112 - 17042790 UGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUU .((((((((.....((.((((....)).)).))....))).)))))(((--((.((((((((((------(....)))))))))((((.......))))....)).)))))......... ( -36.00, z-score = -2.57, R) >droSec1.super_34 161571 101 - 457129 UGUGGACCCAU----------GGAUCAUGACUAACU-GGGAUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUU .((((((((((----------((.......)))..)-))).)))))(((--((.((((((((((------(....)))))))))((((.......))))....)).)))))......... ( -36.20, z-score = -3.30, R) >droYak2.chrX 9258624 112 + 21770863 UAUGGACCCAUCUCUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGACGGCCCGUUUUUAAUACGGGUCCGAAGAGCUGUUAGAAUU ..(((((((.....((.((((....)).)).))....))).)))).(((--.....))).((..------((((((((..((((((((.......))))).)))..)))))))).))... ( -34.80, z-score = -2.36, R) >droEre2.scaffold_4690 12438727 112 + 18748788 UAUGAACUCAUCACUACCCUGAUAUCAUGACUAACUCGAAAUCCAAAGC--UCAUCGCUAUCGA------AGUAGUUUGAUGGCCCGUUCUUAAUACGGGUUCGAAGAGCUGUUAGAAUU (((((....((((......)))).))))).(((((..(.....)..(((--((.((((((((((------(....)))))))))((((.......))))....)).)))))))))).... ( -28.10, z-score = -2.00, R) >droAna3.scaffold_13047 635596 116 - 1816235 UGUGGAGCCCCCA--GCUCUGCCGUCCUGA--GGUUCGGCUUUAAAAGCAGCCAUCUCUGUCGACUGAUGGCUCGUCUGAUGGCCUCUUUUCAAUACGGGUUCGAGAAGCUGUUAGAAUU ...(((((.....--)))))((((..(...--.)..)))).......(.(((((((..........)))))))).(((((((((.((((.((......))...)))).)))))))))... ( -36.10, z-score = -0.27, R) >consensus UGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC__UCAUCGCUAUCGA______AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGUUAGAAUU ..(((((((............................))).)))).........................((((((((..((((((((.......))))).)))..))))))))...... (-19.43 = -19.07 + -0.35)



| Location | 8,790,914 – 8,791,026 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Shannon entropy | 0.38135 |
| G+C content | 0.47931 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.06 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8790914 112 - 22422827 CACGUUUUGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGU .........(((((((.....((.((((....)).)).))....))).)))).(((--((.((((((((((------(....)))))))))((((.......))))....)).))))).. ( -32.60, z-score = -1.60, R) >droSim1.chrX 7008103 112 - 17042790 CACGUUUUGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGU ........((((((((.....((.((((....)).)).))....))).)))))(((--((.((((((((((------(....)))))))))((((.......))))....)).))))).. ( -36.00, z-score = -2.53, R) >droSec1.super_34 161578 101 - 457129 CACGUUUUGUGGACCCAU----------GGAUCAUGACUAACU-GGGAUCCACAGC--UCAUCGCUAUCGA------AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGU ........((((((((((----------((.......)))..)-))).)))))(((--((.((((((((((------(....)))))))))((((.......))))....)).))))).. ( -36.20, z-score = -3.07, R) >droYak2.chrX 9258631 112 + 21770863 CAUCUUUUAUGGACCCAUCUCUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC--UCAUCGCUAUCGA------AGCAGUUUGACGGCCCGUUUUUAAUACGGGUCCGAAGAGCUGU .........(((((((.....((.((((....)).)).))....))).)))).(((--((...(((.....------))).......((((((((.......))))).)))..))))).. ( -33.00, z-score = -2.03, R) >droEre2.scaffold_4690 12438734 112 + 18748788 CACCUUUUAUGAACUCAUCACUACCCUGAUAUCAUGACUAACUCGAAAUCCAAAGC--UCAUCGCUAUCGA------AGUAGUUUGAUGGCCCGUUCUUAAUACGGGUUCGAAGAGCUGU ......((((((....((((......)))).))))))................(((--((.((((((((((------(....)))))))))((((.......))))....)).))))).. ( -26.70, z-score = -1.85, R) >droAna3.scaffold_13047 635603 116 - 1816235 CACCACCUGUGGAGCCCCCA--GCUCUGCCGUCCUGA--GGUUCGGCUUUAAAAGCAGCCAUCUCUGUCGACUGAUGGCUCGUCUGAUGGCCUCUUUUCAAUACGGGUUCGAGAAGCUGU ....(((((((((((.....--)))))((((((..((--.(....(((.....)))(((((((..........)))))))).)).))))))...........))))))............ ( -35.30, z-score = 0.08, R) >consensus CACCUUUUGUGGACCCAUCACUACUCUGAUAUCAUGACUAACUGGGGAUCCAAAGC__UCAUCGCUAUCGA______AGCAGUUUGAUGGCCCGUUUUUAAUACGGGUUCGAAGAGCUGU .........(((((((............................))).))))..........................(((((((..((((((((.......))))).)))..))))))) (-18.27 = -18.06 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:52 2011