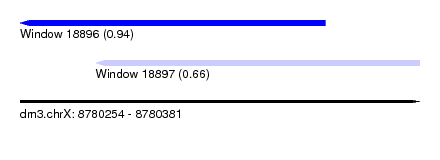
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,780,254 – 8,780,381 |
| Length | 127 |
| Max. P | 0.938417 |

| Location | 8,780,254 – 8,780,351 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.25061 |
| G+C content | 0.51654 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8780254 97 - 22422827 CAGACGACGGGGCGGAUGAGUAACGUGUUAGCAUACUGGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCUCCUAAAGUACCUACCUGAGUGUGU ..(((.(((..((......))..)))))).((((((.((((((....).)))))......(((((((..((((.....))))..))))))))))))) ( -32.70, z-score = -1.30, R) >droSim1.chrX 6997615 92 - 17042790 CAGGCAACGGGGCGGAUGAGUAACGUGUUAGCAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCUC---AAGUACCUACCUGAGUGU-- ((((....(((.((.(((((((..((....)).))))))).)).((((((((((((.....)))))))).).))---)....))).)))).....-- ( -30.40, z-score = -0.87, R) >droSec1.super_34 151341 88 - 457129 CAGGCAACGGGGCGGAUGAAUAACGUGUUAGCAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAAGCUC---AAGUACCUACCUGU------ ......(((((..((.........((....)).((((.(.(((....)((((((((.....)))))))).)).)---.))))))..)))))------ ( -27.00, z-score = -0.82, R) >droYak2.chrX 9247965 92 + 21770863 CGGGCAACGGGGCGGAUGAGUAACGUGUUAACAUACUCAUGCGCUGACUGUACCUGAUUUUCGGGUACAUGCUCA-AGCUCAAGUACCUGAGU---- .(((((....((((.(((((((..((....)).))))))).))))...((((((((.....))))))))))))).-.(((((......)))))---- ( -38.10, z-score = -4.21, R) >droEre2.scaffold_4690 12428456 86 + 18748788 CAGGCAACGGGGCGGAUGAGUAACGUGUUAACAUACUCGUGCGCUGACUGUACCUGAUUUUCGGGUACAUGCUCA-------GGUACCUGAGU---- ..(....)..((((.(((((((..((....)).))))))).))))...((((((((.....)))))))).(((((-------(....))))))---- ( -36.00, z-score = -3.72, R) >consensus CAGGCAACGGGGCGGAUGAGUAACGUGUUAGCAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCUC___AAGUACCUACCUGAGU____ .......((((.((.(((((((..((....)).))))))).)).....((((((((.....)))))))).................))))....... (-25.46 = -25.46 + 0.00)
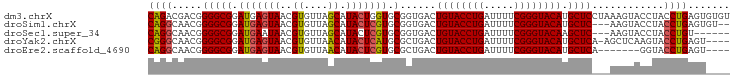
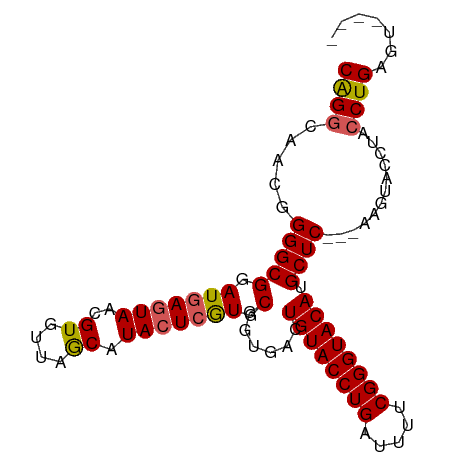
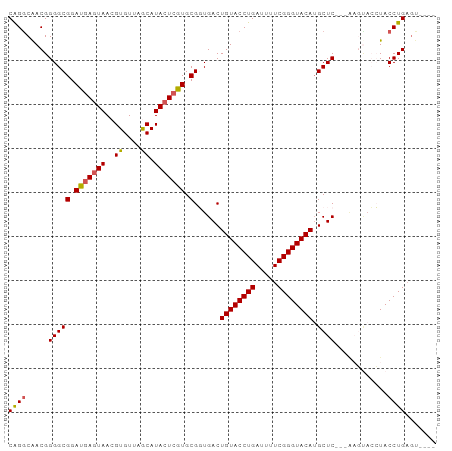
| Location | 8,780,278 – 8,780,381 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.42878 |
| G+C content | 0.49462 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -18.97 |
| Energy contribution | -20.39 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8780278 103 - 22422827 --GCGCAGCUUUGGCCUAAUAAAUAUGAA--ACACAGACGACGGGGCGGAUGAGUAACGUGUUAGCAUACUGGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCU --..(((......(((.....((((((..--((.((..((......))..)).))..)))))).((((....)))))))...((((((((.....))))))))))). ( -29.20, z-score = -0.55, R) >droSim1.chrX 6997634 103 - 17042790 --GCGCAGCUUUGGCCUAAUAAAAAUAAA--ACACAGGCAACGGGGCGGAUGAGUAACGUGUUAGCAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCU --((((.((((((((((............--....))))..))))))....(((((..((....)).)))))))))(....)((((((((.....)))))))).... ( -32.49, z-score = -1.56, R) >droSec1.super_34 151356 103 - 457129 --GCGCAGCUUUGGCCUAAUAAAAAUAAA--ACACAGGCAACGGGGCGGAUGAAUAACGUGUUAGCAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAAGCU --((((.((((((((((............--....))))..))))))...........(((....)))....))))(....)((((((((.....)))))))).... ( -28.89, z-score = -0.90, R) >droYak2.chrX 9247984 105 + 21770863 --GCGCAGCUUUGGCCCAAUAAAACUAAACUAAACGGGCAACGGGGCGGAUGAGUAACGUGUUAACAUACUCAUGCGCUGACUGUACCUGAUUUUCGGGUACAUGCU --((((.((((((((((..................))))..))))))..(((((((..((....)).)))))))))))....((((((((.....)))))))).... ( -36.97, z-score = -3.46, R) >droEre2.scaffold_4690 12428469 103 + 18748788 --GCGCAGCUUUGGCCCAAUAAAACUAAA--AUACAGGCAACGGGGCGGAUGAGUAACGUGUUAACAUACUCGUGCGCUGACUGUACCUGAUUUUCGGGUACAUGCU --(((((((....((((.......((...--....))......))))(.(((((((..((....)).))))))).)))))..((((((((.....))))))))))). ( -34.72, z-score = -2.49, R) >droPer1.super_13 1780479 87 + 2293547 ACGCUUUUUUGUGGCCCAAUAAAU----------CAAGUGCCC--GCGGAUGAGUAA----UUUACAUGC-CAUGUCGCCUUUGCCUCCGGCCUCCGGCCUCUG--- .(((((.(((((......))))).----------.)))))...--(((((((.(((.----......)))-))).))))..........((((...))))....--- ( -17.10, z-score = 1.22, R) >consensus __GCGCAGCUUUGGCCCAAUAAAAAUAAA__ACACAGGCAACGGGGCGGAUGAGUAACGUGUUAACAUACUCGUGCGGUGACUGUACCUGAUUUUCGGGUACAUGCU ....(((((((((((((..................))))..))))))(.(((((((...........))))))).)......(((((((((...)))))))))))). (-18.97 = -20.39 + 1.42)

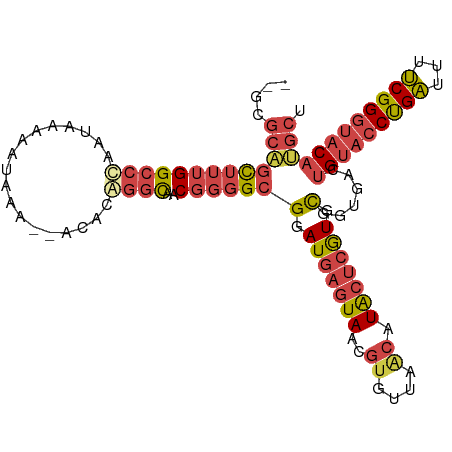
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:49 2011