| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,773,296 – 8,773,369 |
| Length | 73 |
| Max. P | 0.955194 |

| Location | 8,773,296 – 8,773,369 |
|---|---|
| Length | 73 |
| Sequences | 10 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.51369 |
| G+C content | 0.50188 |
| Mean single sequence MFE | -18.93 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.54 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
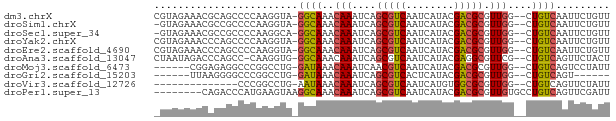
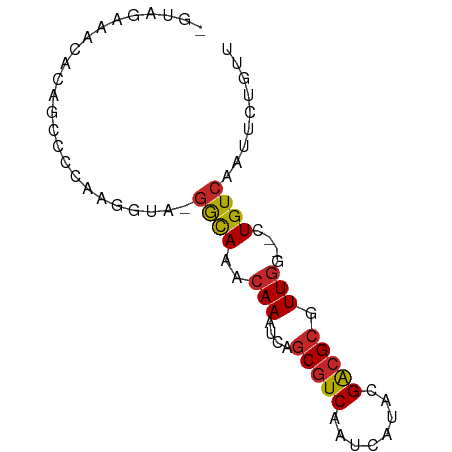
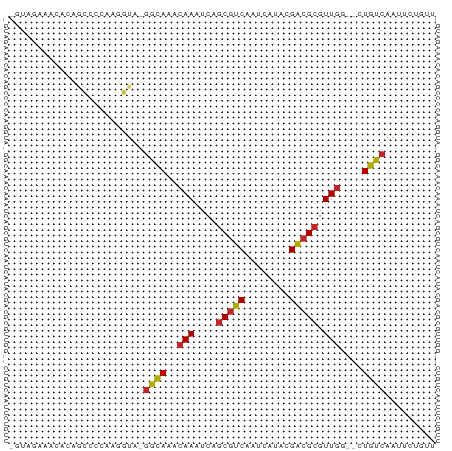
>dm3.chrX 8773296 73 + 22422827 CGUAGAAACGCAGCCCCAAGGUA-GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG--CUGUCAAUUCUGUU ..(((((..((((((((......-))...........(((((........)))))...))--))))...))))).. ( -20.90, z-score = -2.22, R) >droSim1.chrX 6990320 72 + 17042790 -GUAGAAACGCCGCCCCAAGGUA-GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG--CUGUCAAUUCUGUU -.(((((..(((.......))).-((((..(((....(((((........))))).))).--.))))..))))).. ( -20.80, z-score = -2.12, R) >droSec1.super_34 144562 72 + 457129 -GUAGAAACGCCGCCCCAAGGCA-GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG--CUGUCAAUUCUGUU -.(((((..(((.......))).-((((..(((....(((((........))))).))).--.))))..))))).. ( -23.20, z-score = -2.78, R) >droYak2.chrX 9240751 73 - 21770863 CGUAGAAACCCAGCCCCAAGGUA-GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG--CUGUCAAUUCUGUU ..(((((.....(((....))).-((((..(((....(((((........))))).))).--.))))..))))).. ( -19.90, z-score = -2.35, R) >droEre2.scaffold_4690 12421556 73 - 18748788 CGUAGAAACCCAGCCCCAAGGUA-GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG--CUGUCAAUUCUGUU ..(((((.....(((....))).-((((..(((....(((((........))))).))).--.))))..))))).. ( -19.90, z-score = -2.35, R) >droAna3.scaffold_13047 621554 72 + 1816235 CUAAUAGACCCAGCC-CAAGGUG-GGCAAACAAAUCAGCGUCAAUCAUACGAGGCGUUCG--CUGUCAGUUCUACU ....((((....(((-(.....)-)))........(((((....((....))......))--))).....)))).. ( -16.40, z-score = -0.98, R) >droMoj3.scaffold_6473 11079877 67 - 16943266 ------CGGAGAGGCCCGGCCUG-GAUAAACAAAUCAACGUCAAUCAUACGACGCGUUGG--CUGUCAGUCCUAUU ------.(((..(((..((((..-(((......)))..((((........))))....))--)))))..))).... ( -16.10, z-score = -0.10, R) >droGri2.scaffold_15203 6267425 61 + 11997470 ------UUAAGGGGCCCGGCCUG-GAUAAACAAAUCAGCGUCACUCAUACGACGCGUUGG--CUGUCAGU------ ------.....((((...)))).-((((..(((....(((((........))))).))).--.))))...------ ( -17.70, z-score = -0.85, R) >droVir3.scaffold_12726 2623981 59 + 2840439 --------------CCCGGCCUG-AAUAAACAAAUCAGCGUCAAUCAUGUGGCGCGUUGG--CUGUCAGUUCUAUU --------------...(..(((-(.((..(((....((((((......)))))).))).--.))))))..).... ( -16.20, z-score = -1.58, R) >droPer1.super_13 1771455 68 - 2293547 --------CAGACCCAUGAAGUAAGGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGUGCCUGUCAGUUCGAUU --------..((....(((....((((..((((....(((((........))))).)))))))).)))..)).... ( -18.20, z-score = -2.23, R) >consensus _GUAGAAACACAGCCCCAAGGUA_GGCAAACAAAUCAGCGUCAAUCAUACGACGCGUUGG__CUGUCAAUUCUGUU ........................((((..(((....(((((........))))).)))....))))......... (-10.65 = -10.54 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:47 2011