| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,727,109 – 8,727,165 |
| Length | 56 |
| Max. P | 0.534380 |

| Location | 8,727,109 – 8,727,165 |
|---|---|
| Length | 56 |
| Sequences | 8 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 63.54 |
| Shannon entropy | 0.75933 |
| G+C content | 0.47478 |
| Mean single sequence MFE | -13.05 |
| Consensus MFE | -4.91 |
| Energy contribution | -5.62 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
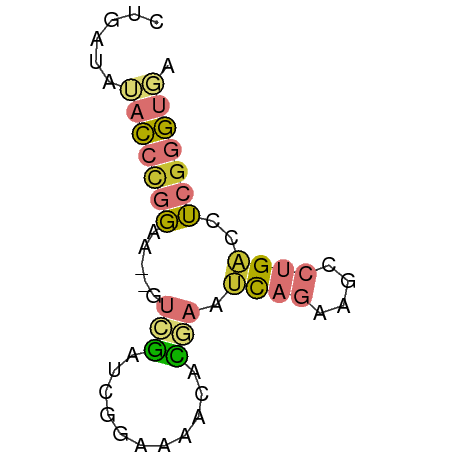
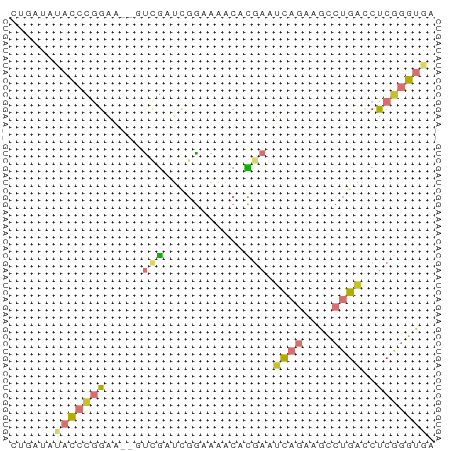
>dm3.chrX 8727109 56 + 22422827 CUGAUAUACCCGGAA--GUCGAUCGGAAAACACGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((..--.....(((.......))).((((....))))..))))))). ( -14.80, z-score = -0.98, R) >droAna3.scaffold_13047 1239411 56 - 1816235 UCCAUGGACCCGGAA--UCGGAAAAGCACUCCCGAAUCAGAAGCCUGACCUCGGGUGA .......((((((..--((((..........)))).((((....))))..)))))).. ( -15.60, z-score = -0.37, R) >droEre2.scaffold_4690 12374474 56 - 18748788 CUGAUGUACCCGGAA--CUCGAUCGGAAAACUCGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((..--.....((((.....)))).((((....))))..))))))). ( -14.60, z-score = -0.67, R) >droYak2.chrX 9191524 58 - 21770863 CCGAUAUACCCGGAAAAGUCGAUCGGAAAACACGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((.........(((.......))).((((....))))..))))))). ( -14.80, z-score = -0.88, R) >droSec1.super_34 100172 56 + 457129 CUGAUAUACCCGGAA--UUCGAUCGGAAAACACGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((..--.....(((.......))).((((....))))..))))))). ( -14.80, z-score = -1.30, R) >droSim1.chrX 6945423 56 + 17042790 CUGAUAUACCCGGAA--GUCGAUCGGAAAACACGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((..--.....(((.......))).((((....))))..))))))). ( -14.80, z-score = -0.98, R) >droWil1.scaffold_181150 3871152 52 - 4952429 AUAAUAGACACCUUGU-AUAGUUUGAAAAACUUCUUCU-----UCUGUCCGCAAGGGU ..........((((((-((((...(((.......))).-----.))))..)))))).. ( -10.40, z-score = -1.50, R) >apiMel3.GroupUn 74603964 55 + 399230636 -UUUCUGUUAUCGUA--UUGCUAUUACAUACUGCACAUACAAAAAAGUUCUAGAAUUA -...........(((--((((...........))).)))).................. ( -4.60, z-score = 0.13, R) >consensus CUGAUAUACCCGGAA__GUCGAUCGGAAAACACGAAUCAGAAGCCUGACCUCGGGUGA ......(((((((.....(((...........))).((((....))))..))))))). ( -4.91 = -5.62 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:43 2011