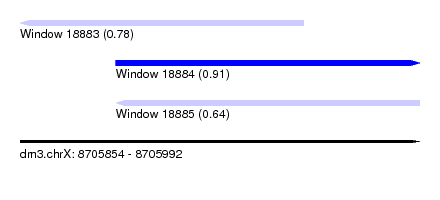
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,705,854 – 8,705,992 |
| Length | 138 |
| Max. P | 0.914157 |

| Location | 8,705,854 – 8,705,952 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.33771 |
| G+C content | 0.41615 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.26 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
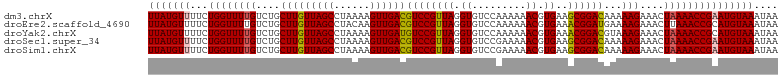
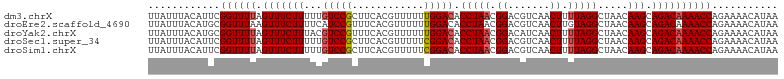
>dm3.chrX 8705854 98 - 22422827 GUUUUUUGGACACCUAAC---GGACGUCAACUUUUAGGCUAACAAGCAGACA---AAACCAGAAAACAUAAAUCUUGGCCACAUUUAUGUGGCUGCAACACCUU (((((((((...((....---))..(((..(.....)(((....))).))).---...))))))))).........(((((((....))))))).......... ( -27.80, z-score = -4.00, R) >droEre2.scaffold_4690 12353895 98 + 18748788 GUUUUUUGGACACCUAAC---GGACGUCAACUUGUAGGCUAACAAGCAGACA---AAACCAGAAAACAUAAAUCUUGGCCACAUUUAGGUGGCAGCAACACCUU (((((((((...((....---))..(((..(((((......)))))..))).---...)))))))))..........(((((......)))))........... ( -30.20, z-score = -4.16, R) >droYak2.chrX 9170339 98 + 21770863 GUUUUUUGGACACCUAAC---GGACAUCAACUUUUAGGCUAACAAGCAGACA---AAACCAGAAAACAUAAAUCUUGGCCACAUUUAGGUGGCUGCAACACCUU (((((((((...((....---))..............(((....))).....---...))))))))).........((((((......)))))).......... ( -25.20, z-score = -3.31, R) >droSec1.super_34 79437 98 - 457129 GUUUUUCGGACACCUAAC---GGACGUCAACUUUUAGGCUAACAAGCAGACA---AAACCAGAAAACAUAAAUCUUGGCCACAUUUAGGUGGCUGCAACACCUU (((((..((...((....---))..(((..(.....)(((....))).))).---...))..))))).........((((((......)))))).......... ( -23.40, z-score = -2.36, R) >droSim1.chrX 6923903 98 - 17042790 GUUUUUCGGACACCUAAC---GGACGUCAACUUUUAGGCUAACAAGCAGACA---AAACCAGAAAACAUAAAUCUUGGCCACAUUUAGGUGGCUGCAACACCUU (((((..((...((....---))..(((..(.....)(((....))).))).---...))..))))).........((((((......)))))).......... ( -23.40, z-score = -2.36, R) >droMoj3.scaffold_6473 10980283 89 + 16943266 -----------CCAUAAA---GGCAGCCAACUUUUAGGCUAACAAGCAGACAGCCGGAAAACCAAACAUAAAUCUU-GUUACAUUUAUGUGGCUCUAACACCUU -----------.....((---((.((((........))))...........((((((....))..((((((((...-.....))))))))))))......)))) ( -18.80, z-score = -1.65, R) >droVir3.scaffold_12726 2535876 92 - 2840439 -----------ACAUAACGAUGGCAGCCAACUUUUAGGCUAACAAGCAGACAGCCGGAAAACCAAACAUAAAUCUU-GUUACAUUUAUGUGGCUCUAACACCUU -----------.........((..((((........))))..)).......((((((....))..((((((((...-.....)))))))))))).......... ( -18.50, z-score = -1.49, R) >droGri2.scaffold_14853 8688449 88 - 10151454 -----------ACAUAAC---GGCAGCCAACUUUUAGGCUAACAAGCAGACAGCCGGAAAACCAAACAUAAAUCUU-GUCACAUUUAUGUGGCUCUAGUACCU- -----------.......---(..((((........))))..)..(((((..(((((....))..((((((((...-.....)))))))))))))).))....- ( -18.30, z-score = -1.28, R) >consensus GUUUUU_GGACACCUAAC___GGACGUCAACUUUUAGGCUAACAAGCAGACA___AAACCAGAAAACAUAAAUCUUGGCCACAUUUAGGUGGCUGCAACACCUU .....................((..............(((....)))..............................(((((......))))).......)).. (-10.68 = -10.26 + -0.42)

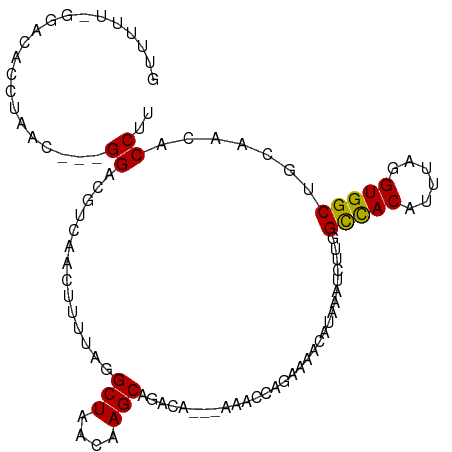
| Location | 8,705,887 – 8,705,992 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Shannon entropy | 0.07137 |
| G+C content | 0.36762 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8705887 105 + 22422827 UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUAAAAGUUGACGUCCGUUAGGUGUCCAAAAAACGUGAAGCGGACAAAAAGAAACUAAAACCGAAUGUAAAUAA ((((((((...(((((((....(((((((((......))))))((((((((.((.........)).))..))))))...)))....))))))))))))))).... ( -24.80, z-score = -2.31, R) >droEre2.scaffold_4690 12353928 105 - 18748788 UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUACAAGUUGACGUCCGUUAGGUGUCCAAAAAACGUGAAACGGAUGAAAAGAAACUUAAACCGCAUGUAAAUAA .(((((.....(((((((((.((((((......)))))).)))(((((((..((((.......))))..))))))).....))))))......)))))....... ( -24.50, z-score = -1.70, R) >droYak2.chrX 9170372 105 - 21770863 UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUAAAAGUUGAUGUCCGUUAGGUGUCCAAAAAACGUGAAACGGACGUAAAGAAACUAAAACCGCAUGUAAAUAA .(((((....(((((((...(((..((((((......))))))(((((((..((((.......))))..))))))))))..))))))).....)))))....... ( -21.30, z-score = -1.11, R) >droSec1.super_34 79470 105 + 457129 UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUAAAAGUUGACGUCCGUUAGGUGUCCGAAAAACGUGAAGCGGACAAAAAGAAACUAAAACCGAAUGUAAAUAA ((((((((...(((((((....(((((((((......))))))(((((((.....((((.....)).)))))))))...)))....))))))))))))))).... ( -24.80, z-score = -2.09, R) >droSim1.chrX 6923936 105 + 17042790 UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUAAAAGUUGACGUCCGUUAGGUGUCCGAAAAACGUGAAGCGGACAAAAAGAAACUAAAACCGAAUGUAAAUAA ((((((((...(((((((....(((((((((......))))))(((((((.....((((.....)).)))))))))...)))....))))))))))))))).... ( -24.80, z-score = -2.09, R) >consensus UUAUGUUUUCUGGUUUUGUCUGCUUGUUAGCCUAAAAGUUGACGUCCGUUAGGUGUCCAAAAAACGUGAAGCGGACAAAAAGAAACUAAAACCGAAUGUAAAUAA (((((((...((((((((....(((((((((......))))))((((((((.((.........)).))..))))))...)))....))))))))))))))).... (-22.56 = -22.60 + 0.04)
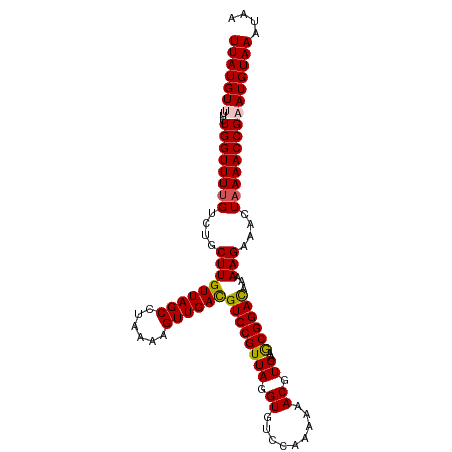
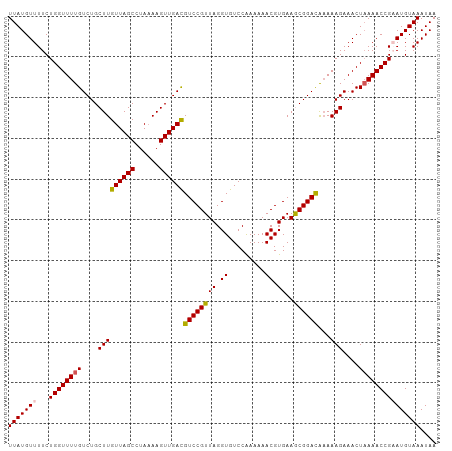
| Location | 8,705,887 – 8,705,992 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.81 |
| Shannon entropy | 0.07137 |
| G+C content | 0.36762 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.636970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8705887 105 - 22422827 UUAUUUACAUUCGGUUUUAGUUUCUUUUUGUCCGCUUCACGUUUUUUGGACACCUAACGGACGUCAACUUUUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA .........(((((((((.(((((((.(((.((.....(((((..((((....))))..))))).........)).))).))).)))))))))).)))....... ( -19.94, z-score = -1.67, R) >droEre2.scaffold_4690 12353928 105 + 18748788 UUAUUUACAUGCGGUUUAAGUUUCUUUUCAUCCGUUUCACGUUUUUUGGACACCUAACGGACGUCAACUUGUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA ..........((((...(((.....)))...)))).....(((((((((...((....))..(((..(((((......)))))..)))....))))))))).... ( -24.80, z-score = -2.88, R) >droYak2.chrX 9170372 105 + 21770863 UUAUUUACAUGCGGUUUUAGUUUCUUUACGUCCGUUUCACGUUUUUUGGACACCUAACGGACAUCAACUUUUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA ............((((((.((((......(((((((....(((.....)))....)))))))............(((....)))))))))))))........... ( -21.60, z-score = -2.50, R) >droSec1.super_34 79470 105 - 457129 UUAUUUACAUUCGGUUUUAGUUUCUUUUUGUCCGCUUCACGUUUUUCGGACACCUAACGGACGUCAACUUUUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA .........(((((((((.(((((((..((((((............))))))(((((.((.......)).))))).....))).)))))))))).)))....... ( -20.50, z-score = -1.98, R) >droSim1.chrX 6923936 105 - 17042790 UUAUUUACAUUCGGUUUUAGUUUCUUUUUGUCCGCUUCACGUUUUUCGGACACCUAACGGACGUCAACUUUUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA .........(((((((((.(((((((..((((((............))))))(((((.((.......)).))))).....))).)))))))))).)))....... ( -20.50, z-score = -1.98, R) >consensus UUAUUUACAUUCGGUUUUAGUUUCUUUUUGUCCGCUUCACGUUUUUUGGACACCUAACGGACGUCAACUUUUAGGCUAACAAGCAGACAAAACCAGAAAACAUAA ............((((((.(((((((...(((((............))))).(((((.((.......)).))))).....))).))))))))))........... (-17.02 = -17.38 + 0.36)
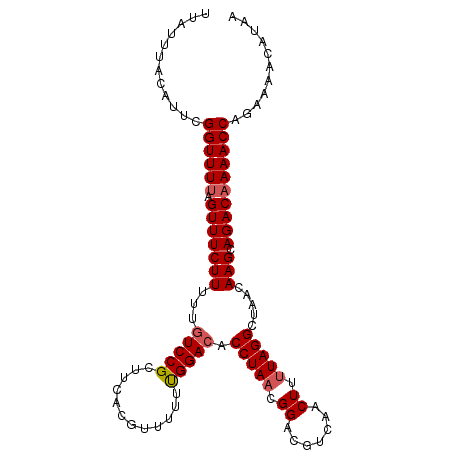
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:39 2011