| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,685,794 – 8,685,895 |
| Length | 101 |
| Max. P | 0.733836 |

| Location | 8,685,794 – 8,685,895 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Shannon entropy | 0.60041 |
| G+C content | 0.48770 |
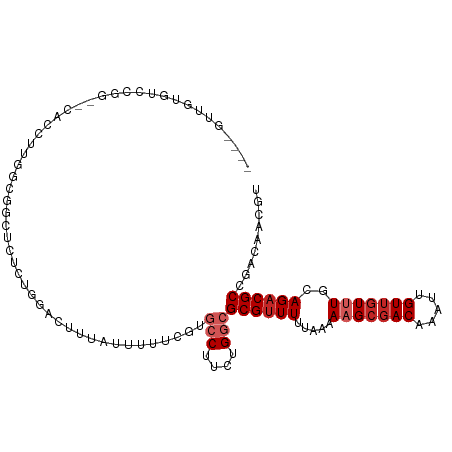
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.00 |
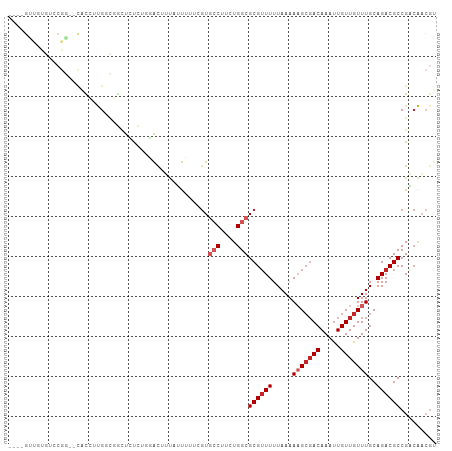
| Mean z-score | -1.37 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733836 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
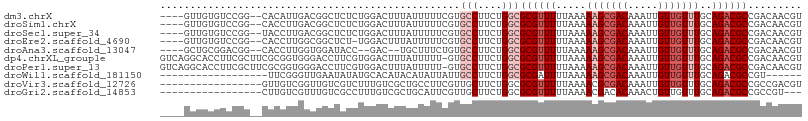
>dm3.chrX 8685794 101 + 22422827 ----GUUGUGUCCGG--CACAUUGACGGCUCUCUGGACUUUAUUUUUCGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ----..((((.....--))))(((.((((..((((...((((.....((((((....))))))....))))(((((((.....))))))).)))).)))).)))... ( -32.00, z-score = -1.84, R) >droSim1.chrX 6903848 101 + 17042790 ----GUUGUGUCCGG--CACCUUGACGGCUCUCUGGACUUUAUUUUUCGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ----(((((((((((--..((.....))....))))))((((.....((((((....))))))....))))..))))).....((((((.((....)).)))))).. ( -33.00, z-score = -2.19, R) >droSec1.super_34 59730 101 + 457129 ----GUUGUGUCCGG--UACCUUGACGGCUCUCUGGACUUUAUUUUUCGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ----(((((((((((--..((.....))....))))))((((.....((((((....))))))....))))..))))).....((((((.((....)).)))))).. ( -33.70, z-score = -2.69, R) >droEre2.scaffold_4690 12334360 100 - 18748788 ----GUUGUGUCCGG--CACCUUGGCGGCUCU-UGGACUUUAUUUUUCGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ----(((((((((((--..((.....))...)-)))))((((.....((((((....))))))....))))..))))).....((((((.((....)).)))))).. ( -31.40, z-score = -1.23, R) >droAna3.scaffold_13047 538872 97 + 1816235 ----GCUGCGGACGG--CACCUUGGUGGAUACC--GAC--UGCUUUCUGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ----...(((..(((--(...(((((....)))--))(--(((.....(((((....))))).........(((((((.....)))))))))))..))))....))) ( -32.20, z-score = -1.33, R) >dp4.chrXL_group1e 7958938 106 + 12523060 GUCAGGCACCUUCGCUUCGCGGUGGGACCUUCGUGGACUUUAUUUUU-GUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ((((((..((.((((...)))).))..)))..((....((((....(-(((((....))))))....))))..))))).....((((((.((....)).)))))).. ( -32.60, z-score = -0.79, R) >droPer1.super_13 1687210 106 - 2293547 GUCAGGCACCUUCGCUUCGCGGUGGGACCUUCGUGGACUUUAUUUUU-GUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ((((((..((.((((...)))).))..)))..((....((((....(-(((((....))))))....))))..))))).....((((((.((....)).)))))).. ( -32.60, z-score = -0.79, R) >droWil1.scaffold_181150 3807175 83 - 4952429 ------------------UUCGGGUUGAAUAUAUGCACAUACAUAUUAUUGCCUUCUGGCGCGAUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGU------ ------------------...((((.....(((((......)))))....))))...(((((.........(((((((.....)))))))...).))))..------ ( -17.50, z-score = 0.12, R) >droVir3.scaffold_12726 2507601 90 + 2840439 -----------------GUUGUCGGUUGUCGUCUUUGUCGCUGCCUUCGUUGCUUCUGGCGCGUUUUUAAAACGCGACAAAUUGUUGUUUGCAGACGCCGCCGACGU -----------------...((((((.(.((((((((((...(((............)))(((((.....))))))))))).(((.....)))))))).)))))).. ( -32.50, z-score = -2.22, R) >droGri2.scaffold_14853 8661197 87 + 10151454 -----------------CUUGUCGUUUGUCGCCUUUGUCGCUGCAUUCGUUGCUUCUGGCGCGUUUUUAAAACGACACAAACUGUUGUUUGCAGACGCCGCCGU--- -----------------.....((..(((.((.......)).)))..))........(((((((((...((((((((.....))))))))..))))).))))..--- ( -24.90, z-score = -0.78, R) >consensus ____GUUGUGUCCGG__CACCUUGGCGGCUCUCUGGACUUUAUUUUUCGUGCCUUCUGGCGCGUUUUUAAAAAGCGACAAAUUGUUGUUUGCAGACGCCGACAACGU ..................................................(((....)))((((((.....(((((((.....)))))))..))))))......... (-13.05 = -13.95 + 0.90)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:34 2011