| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,670,888 – 10,670,999 |
| Length | 111 |
| Max. P | 0.956349 |

| Location | 10,670,888 – 10,670,999 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 74.62 |
| Shannon entropy | 0.52930 |
| G+C content | 0.56189 |
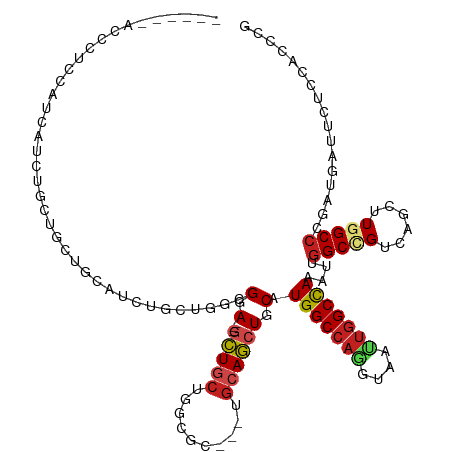
| Mean single sequence MFE | -45.11 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10670888 111 - 23011544 ------ACCCUCCAUCAUUUGCUGCUGCAUCUGUUGGCGGA---GCUGCUGGCGC---UGCAGCUGCAUGGCCAGGUAAUUGGCCAAAUUGGCCGUGAGCUUGGCCCGAUGAUUCUCCACCCG ------......((((...(((.((((((..(((..(((..---..)))..))).---)))))).))).((((((((...(((((.....)))))...)))))))).))))............ ( -50.20, z-score = -2.84, R) >droSim1.chr2L 10468874 111 - 22036055 ------ACCCUCCAUCAUCUGCUGCUGCAUCUGCUGGCGGA---GCUGCUGGCGC---UGCAGCUGCAUGGCCAGGUAAUUGGCCAAAUUGGCCGUGAGCUUGGCCCGAUGAUUCUCCACCCG ------.......(((((((((.((((((..(((..(((..---..)))..))).---)))))).))).((((((((...(((((.....)))))...)))))))).)))))).......... ( -53.20, z-score = -3.38, R) >droSec1.super_3 6088606 111 - 7220098 ------ACCCUCCAACAUCUGCUGCUGCAUCUGCUGGCGGA---GCUGCUGGCGC---UGCAGCUGCAUGGCCAGGUAAUUGGCCAAAUUGGCCGUGAGCUUGGCCCGAUGAUUCUCCACCCG ------.........(((((((.((((((..(((..(((..---..)))..))).---)))))).))).((((((((...(((((.....)))))...)))))))).))))............ ( -51.40, z-score = -2.99, R) >droYak2.chr2L 7075155 111 - 22324452 ------UCCCUCCAUCAUCUGCUGCUGCAUCUGCUGGCGGA---GCUGCUGGCGC---UGCAGCUGCAUGGCCAGAUAAUUGGCCAAAUUGGCCGUAAGCUUGGCCCGAUGAUUCUCCACCCG ------.......(((((((((.((((((..(((..(((..---..)))..))).---)))))).))).((((((.....(((((.....))))).....)))))).)))))).......... ( -48.10, z-score = -2.62, R) >droEre2.scaffold_4929 11879353 111 + 26641161 ------ACCCUCCAUCAUCUGCUGCUGCAUCUGCUGGCGGA---GCUGCUGGCGC---UGCAGCUGCAUGGCCAGAUAAUUGGCCAAAUUGGCCGUGAGCUUGGCCCGAUGAUUCUCCACCCG ------.......(((((((((.((((((..(((..(((..---..)))..))).---)))))).))).((((((.....(((((.....))))).....)))))).)))))).......... ( -48.10, z-score = -2.41, R) >droAna3.scaffold_12916 14514324 111 - 16180835 ------GCCUUCCAUCAUUUGUUGCUGCAUCUGCUGGCGCA---GCUGCUGACGU---GUAAGCUGCAUGGCCAGAUAGUUGGCCAGGUUGGCAGUUAACUUGGCCCGAUGGUUCUCCACCCG ------.....(((((..............(((((((((((---(((((......---)).))))))...))))).)))..((((((((((.....)))))))))).)))))........... ( -40.70, z-score = -0.65, R) >dp4.chr4_group4 2869915 108 - 6586962 ---------CUGCGACAUCUGCAACUGGAUUUGGUUCUGCA---GCUGCUGCCGC---UGCAGCUGCAUGGCCAAGAAGCUGGCCAGAUUGGCCGUCAGCUUUGCCUUGUGGUUCUCGACCCG ---------...(((...(..(((..((..(((((..((((---(((((......---.)))))))))..)))))((((((((((.....))))...)))))).)))))..)...)))..... ( -46.40, z-score = -1.99, R) >droPer1.super_10 1882107 108 - 3432795 ---------CUGCGACAUCUGCAAUUGGAUUUGGUUCUGCA---GCUGCUGCCGC---UGCAGCUGCAUGGCCAAGAAGCUGGCCAGAUUGGCCGUCAGCUUUGCCUUGUGGUUCUCGACCCG ---------...(((...(..(((..((..(((((..((((---(((((......---.)))))))))..)))))((((((((((.....))))...)))))).)))))..)...)))..... ( -46.40, z-score = -2.08, R) >droWil1.scaffold_180708 1891064 102 + 12563649 ---------------------AUUUUGUAUUUGAUUUUGUAUUGUUUGCUGACGCUUUUGGAAUUGCAUGGCCAAGUAUUGUGCUAAAUUUGCCGUCAAUUUGGCCUUAUGAUUCUCCACUCU ---------------------.....(((..((((.....))))..))).........((((....(((((((((((..((.((.......))))...))))))))..)))....)))).... ( -19.80, z-score = -0.63, R) >droVir3.scaffold_12963 9888217 117 + 20206255 AGCUGAUUGAAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGA---GCUGCUGACGC---UGCAGCUGCAUGGCCAGGAAGCUGGCCAAAUUGGCCGUCAGCUUGGCCCUGUGAUUCUCUACGCG ...........((((.(((.(((..............((.(---(((((......---.)))))).)).(((((((..(.(((((.....))))).)..))))))).)))))).....)))). ( -41.70, z-score = -0.49, R) >droMoj3.scaffold_6500 13343377 120 + 32352404 GGCUGAGUGUAGUUGAGUCUGCAUUUGAAUUUGAUUCUGGAGCUGCUGCUGACGC---UGCAGCUGCAUGGCCAGGAAGCUGGCCAAAUUGGCUGUCAGCUUGGCCCGUUGAUUCUCCACGCG .((.((((((((......))))))))...........(((((..((.((..(.((---((((((((..(((((((....)))))))...)))))).)))))..))..)).....))))).)). ( -47.80, z-score = -1.43, R) >droGri2.scaffold_15252 8793661 117 + 17193109 AGCUGAUUGGAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGA---ACUGCUGGCGC---UGCAGCUGCAUGGCCAGGAAACUGGCCAAAUUGGCUGUCAACUUGGCCCUGUGGUUCUCAACGCG ...........((((((..(((....))).........(((---((..(.((.((---((((((((..(((((((....)))))))...)))))))......))))).)..))))))))))). ( -47.50, z-score = -2.78, R) >consensus ______ACCCUCCAUCAUCUGCUGCUGCAUCUGCUGGCGGA___GCUGCUGGCGC___UGCAGCUGCAUGGCCAGGUAAUUGGCCAAAUUGGCCGUCAGCUUGGCCCGAUGAUUCUCCACCCG ...................(((....((........))......(((((..........))))).)))(((((((....)))))))....(((((......)))))................. (-21.69 = -22.33 + 0.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:41 2011