| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,666,739 – 8,666,835 |
| Length | 96 |
| Max. P | 0.649530 |

| Location | 8,666,739 – 8,666,835 |
|---|---|
| Length | 96 |
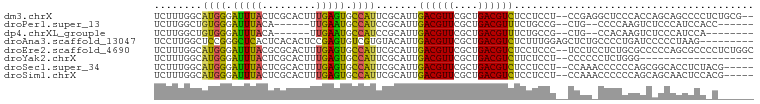
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.65781 |
| G+C content | 0.56273 |
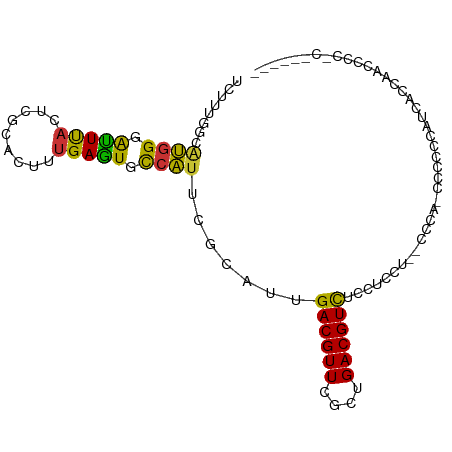
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.81 |
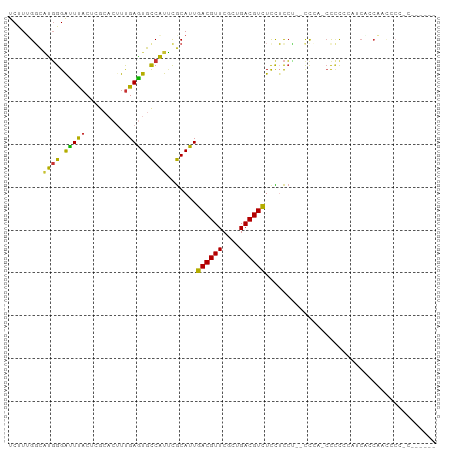
| Combinations/Pair | 1.53 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.649530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8666739 96 + 22422827 UCUUUGGCAUGGGAUUUACUCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUCCUCCU--CCGAGGCUCCCACCAGCAGCCCCUCUGCG-- ....(((..(((((....(((((((.....))))..........((((((....)))))).......--..)))..))))))))((((.....)))).-- ( -28.40, z-score = -1.39, R) >droPer1.super_13 1666932 84 - 2293547 UCUUGGCUGUGGGAUUUACA------UUGAAUGCCAUCCGCAUUGACGUUCGCUGACGUUUCUGCCG--CUG--CCCCAAGUCUCCCAUCCACC------ ...(((..((((((...((.------(((...((...(.(((..((((((....))))))..))).)--..)--)..))))).)))))))))..------ ( -20.30, z-score = -0.66, R) >dp4.chrXL_group1e 7938863 82 + 12523060 UCUUGGCUGUGGGAUUUACA------UUGAAUGCCAUCCGCAUUGACGUUCGCUGACGUUUCUGCCG--CUG--CCACAAGUCUCCCAUCCA-------- ...((((.(((((((...((------(...)))..))))(((..((((((....))))))..)))))--).)--)))...............-------- ( -20.80, z-score = -0.65, R) >droAna3.scaffold_13047 522242 91 + 1816235 UCCUUGGCUCCGGGCUCACUCACACUCCGAGUGUCGUGUACAUUGACGUUCGCUGACGUCUCUUUGGAGCUCUGCCCCUGAUCCCCCUAAG--------- .....(((...((((((((.(((((.....)))).).)).((..((((((....))))))....)))))))).)))...............--------- ( -24.50, z-score = -0.77, R) >droEre2.scaffold_4690 12316080 98 - 18748788 UCUUUGGCAUGGGAUUUACGCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUCCUCCC--UCCUCCUCUGCGCCCCCAGCGCCCCUCUGGC .....(((.((((......(((((.....(((((.....)))))((((((....)))))).......--........))))).))))..)))........ ( -26.40, z-score = -0.63, R) >droYak2.chrX 9130116 79 - 21770863 UCUUUGGCAUGGGAUUUACUCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUUCUCCU--CCCCCCUCUGGG------------------- .((..((...((((.......((((.....))))..........((((((....))))))......)--))).))...)).------------------- ( -18.10, z-score = -0.23, R) >droSec1.super_34 40965 93 + 457129 UCUUUGGCAUGGGAUUUACUCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUCCUCCU--CCAAACCCCCCAGCGGCACCUCUACG----- ......((.((((........((((.....))))..........((((((....)))))).......--........))))...)).........----- ( -20.40, z-score = -0.45, R) >droSim1.chrX 6891705 93 + 17042790 UCUUUGGCAUGGGAUUUACUCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUCCUCCU--CCAAACCCCCCAGCAGCAACUCCACG----- ......((.((((........((((.....))))..........((((((....)))))).......--........))))))............----- ( -19.90, z-score = -0.64, R) >consensus UCUUUGGCAUGGGAUUUACUCGCACUUUGAGUGCCAUUCGCAUUGACGUUCGCUGACGUCUCCUCCU__CCCA_CCCCCCAUCACCAACCCC_C______ ........((((.(((((.........))))).)))).......((((((....))))))........................................ (-11.21 = -10.40 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:31 2011