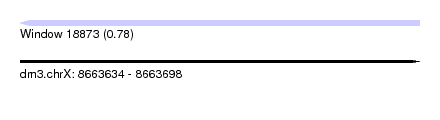
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,663,634 – 8,663,698 |
| Length | 64 |
| Max. P | 0.777604 |

| Location | 8,663,634 – 8,663,698 |
|---|---|
| Length | 64 |
| Sequences | 12 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 90.57 |
| Shannon entropy | 0.19468 |
| G+C content | 0.37971 |
| Mean single sequence MFE | -15.72 |
| Consensus MFE | -13.31 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777604 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 8663634 64 - 22422827 UGGGUUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- ..((((((..(....)((((.((((((((((........))))))))))..)))-)))))))...- ( -16.00, z-score = -2.05, R) >droSim1.chrX 6889343 64 - 17042790 UGGGUUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- ..((((((..(....)((((.((((((((((........))))))))))..)))-)))))))...- ( -16.00, z-score = -2.05, R) >droSec1.super_34 38014 64 - 457129 UGGGUUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- ..((((((..(....)((((.((((((((((........))))))))))..)))-)))))))...- ( -16.00, z-score = -2.05, R) >droYak2.chrX 9127116 64 + 21770863 UGGGUUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- ..((((((..(....)((((.((((((((((........))))))))))..)))-)))))))...- ( -16.00, z-score = -2.05, R) >droEre2.scaffold_4690 12313332 64 + 18748788 UGGGUUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- ..((((((..(....)((((.((((((((((........))))))))))..)))-)))))))...- ( -16.00, z-score = -2.05, R) >droAna3.scaffold_13047 519543 62 - 1816235 --GGCUGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU-GGCAACUGUC- --.(((((((...))))(((.((((((((((........)))))))))).))).-))).......- ( -14.50, z-score = -1.29, R) >dp4.chrXL_group1e 7936304 64 - 12523060 UUGGUCGCGUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUUU-GGCAAGUGCC- ..((.(((......(((.((.((((((((((........))))))))))...))-.))).)))))- ( -14.70, z-score = -1.11, R) >droPer1.super_13 1664354 64 + 2293547 UUGGUCGCGUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUUU-GGCAAGUGCC- ..((.(((......(((.((.((((((((((........))))))))))...))-.))).)))))- ( -14.70, z-score = -1.11, R) >droWil1.scaffold_181150 3783118 64 + 4952429 UGAGCCGCAUCUGGUGCGCAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUU--GGCAAGUUGUU ...(((((((...))))....((((((((((........))))))))))....--)))........ ( -14.70, z-score = -0.69, R) >droVir3.scaffold_12726 2480172 65 - 2840439 UGUGCCGCAUCUGCUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUUUCGGCAAGUCUC- ..(((((((.....)))(((.((((((((((........))))))))))...)))))))......- ( -17.00, z-score = -1.90, R) >droMoj3.scaffold_6473 10923508 65 + 16943266 UGGGCCGCAUCUGCUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUUUCGGCAAGUCUC- ...((((((.....)))(((.((((((((((........))))))))))...)))))).......- ( -16.10, z-score = -1.34, R) >droGri2.scaffold_14853 8633249 65 - 10151454 UGUGCCGCAUCUGCUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUUUCGGCAACUCUU- ..(((((((.....)))(((.((((((((((........))))))))))...)))))))......- ( -17.00, z-score = -2.02, R) >consensus UGGGUCGCAUCUGGUGCGGAUUAGAUUAUGCAUAAUAAAGUGUAAUUUAUUUCU_GGCAACUGUC_ ...((((((.....)))(((.((((((((((........)))))))))).)))..)))........ (-13.31 = -12.52 + -0.80)

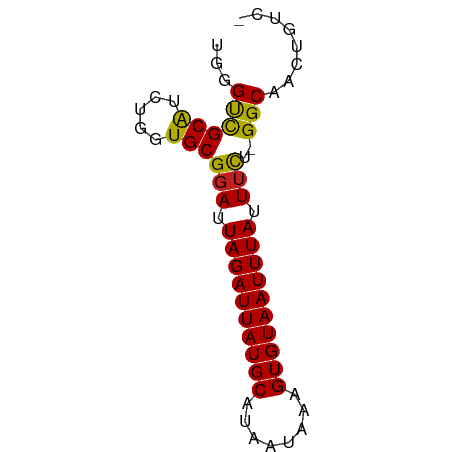

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:30 2011