
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,647,778 – 8,647,876 |
| Length | 98 |
| Max. P | 0.526925 |

| Location | 8,647,778 – 8,647,876 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.51 |
| Shannon entropy | 0.70715 |
| G+C content | 0.38478 |
| Mean single sequence MFE | -19.29 |
| Consensus MFE | -5.32 |
| Energy contribution | -5.62 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
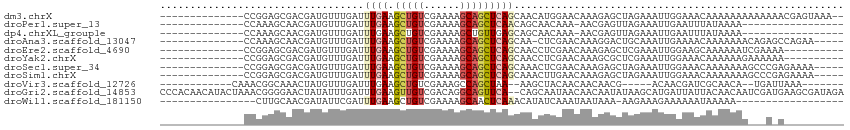
>dm3.chrX 8647778 98 - 22422827 --------------CCGGAGCGACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAACAUGGAACAAAGAGCUAGAAAUUGGAAACAAAAAAAAAAAAACGAGUAAA-- --------------.((.(((..(..(((((.(((((.((((((......))))))...))).)).)))))..).))).....(((....)))...........))......-- ( -21.10, z-score = -2.06, R) >droPer1.super_13 1645988 82 + 2293547 --------------CCAAAGCAACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAACAGCAACAAA-AACGAGUUAGAAAUUGAAUUUAUAAAA----------------- --------------.........((((.(((((((((..(((.......))).(((....)))......-..))))))))).))))...........----------------- ( -14.40, z-score = -0.99, R) >dp4.chrXL_group1e 7919434 82 - 12523060 --------------CCAAAGCAACGAUGUUUGAUUUGAAGCUGUCGAAAAGCUGUUGAGCAGCAACAAA-AACGAGUUAGAAAUUGAAUUUAUAAAA----------------- --------------.........((((.(((((((((.((((.......))))((((.....))))...-..))))))))).))))...........----------------- ( -15.80, z-score = -1.18, R) >droAna3.scaffold_13047 502532 94 - 1816235 --------------CCAAAGCAACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAA-CUCGAACAAAGGACUGCAAAUUGAAAACAAAAAAACAGAGCCAGAA----- --------------.....(((.(..(((((((.(((.((((((......))))))...)))-.)))))))..)...))).............................----- ( -20.40, z-score = -1.48, R) >droEre2.scaffold_4690 12295858 90 + 18748788 --------------CCGGAGCGACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAACCUCGAACAAAGAGCUCGAAAUUGGAAGCAAAAAAUCGAAAA---------- --------------.((((((..(..(((((((.(((.((((((......))))))...)))..)))))))..).))))....(((....))).....))....---------- ( -23.20, z-score = -1.21, R) >droYak2.chrX 9110513 90 + 21770863 --------------CCGGAGCGACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAACCUCGAACAAAGCGCUCGAAAUUGGAAACAAAAAAGAAAAAA---------- --------------...(((((.(..(((((((.(((.((((((......))))))...)))..)))))))..))))))....(((....)))...........---------- ( -28.30, z-score = -3.52, R) >droSec1.super_34 22711 95 - 457129 --------------CCGGAGCGACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAAACUCGAACAAAGAGCUAGAAAUUGGAAACAAAAAAAGCCCGAGAAAA----- --------------.(((.((.....(((((((((((.((((((......))))))...)))).)))))))............(((....))).....)))))......----- ( -27.50, z-score = -3.48, R) >droSim1.chrX 6874296 95 - 17042790 --------------CCGGAGCGACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAAACUUGAACAAAGAGCUAGAAAUUGGAAACAAAAAAAGCCCGAGAAAA----- --------------.(((.((.....((((..(((((.((((((......))))))...)))).)..))))............(((....))).....)))))......----- ( -25.30, z-score = -2.99, R) >droVir3.scaffold_12726 2459945 86 - 2840439 ------------CAAACGGCAAACUAUGUUUGAUUUGAAGCUGUCGAAAGCCAGCUAA--AAGCUACAACAACAACG-----ACAACGAUCGCAACA--UGAUUAAA------- ------------((((((........))))))......(((((.(....).)))))..--.................-----.....(((((.....--)))))...------- ( -14.50, z-score = -0.70, R) >droGri2.scaffold_14853 8611826 112 - 10151454 CCCACAACAUACUAAACGGGGAACUAUAUUUGAUUUGAAGUUGUCGACAGGCAGUUCA--CAGCAAUAACAACAAUAUAAGCAUGAUUAUUACAACAAUCGAUGAAGCGAUAGA (((..............)))...((((..(((...((((.(((((....)))))))))--...)))...............(((((((........)))).))).....)))). ( -14.54, z-score = 0.36, R) >droWil1.scaffold_181150 3758723 79 + 4952429 ----------------CUUGCAACGAUAUUCGAUUUGAAGCUGUCGAAAAGCAACUCAAACAUAUCAAAUAAUAAA-AAGAAAGAAAAAAUAAAAA------------------ ----------------........(((((....(((((.(((.......)))...))))).)))))..........-...................------------------ ( -7.10, z-score = -0.41, R) >consensus ______________CCGGAGCAACGAUGUUUGAUUUGAAGCUGUCGAAAAGCAGCUCAGCAACCUCGAACAAAGAGCUAGAAAUUGGAAACAAAAAAA___A_A__________ ..................................((((.(((((......)))))))))....................................................... ( -5.32 = -5.62 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:29 2011