| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,641,970 – 8,642,085 |
| Length | 115 |
| Max. P | 0.974867 |

| Location | 8,641,970 – 8,642,085 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.68 |
| Shannon entropy | 0.70694 |
| G+C content | 0.46647 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
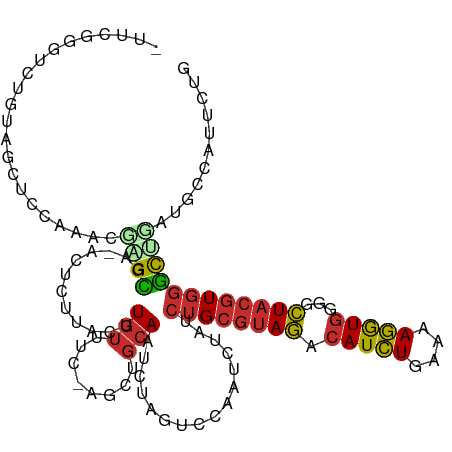
>dm3.chrX 8641970 115 + 22422827 UUUCGGGUCUGUAGCUCCAAACGAGCGA-ACUCUUACUUGUUUC-AGCUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCCGAUGCCAUUCUG ..((((.(((((((((..(((((((..(-....)..))))))).-)))))))((.((((((..(((((....)))))(((((....))))))))))).)))).)))).......... ( -37.90, z-score = -2.25, R) >droSim1.chrX_random 2584926 115 + 5698898 UGUCGGGUCUGUAGCUUCAAACGAGCGA-AUUCUUACUUGUUUC-AGCUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCCGAUGCCAUUCUG .(((((.(((((((((..(((((((..(-....)..))))))).-)))))))((.((((((..(((((....)))))(((((....))))))))))).)))).)))))......... ( -40.00, z-score = -2.89, R) >droSec1.super_34 18705 115 + 457129 UUUCGGGUCUGUAGCUCCAAACGAGCGA-AUUCUUACUUGUUUC-AGCUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCCGAUGCCAUUCUG ..((((.(((((((((..(((((((..(-....)..))))))).-)))))))((.((((((..(((((....)))))(((((....))))))))))).)))).)))).......... ( -37.90, z-score = -2.36, R) >droYak2.chrX 9107009 115 - 21770863 UUUGGGGUCUGUAGCUCCAAAGCAGCGA-AUUCUUACUUGUUUC-AGCUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCUGAUGCUAUUCUG ((((((((.....))))))))(((((((-(..(......).)))-.)))))....((((...(((..((..(((((.(((((....)))))...)))))..))..)))))))..... ( -39.90, z-score = -2.78, R) >droEre2.scaffold_4690 12291916 113 - 18748788 -UUUGGGUCUGUAGCACGAAAGCAGCGA-AUCCCUACUUGUUUC-A-CUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCUGAUGCUAUUCUG -....((...(((((......((((.((-(...........)))-.-))))...........(((..((..(((((.(((((....)))))...)))))..))..)))))))).)). ( -34.60, z-score = -1.48, R) >droAna3.scaffold_13047 498098 114 + 1816235 CACCUAGUCGAUA-CUAAUUACCGGCAG-CCUGGCUUAUUUUUU-AGUUGCAAUCUAGAGCAAUUUAUCUGCGCAGACAUCUGAAAAGGUGGGGCUACGUGGGCUGCUGCGAGUCUG ....((((....)-)))....(((((((-(((.((..((.....-((((((........)))))).))..))((...(((((....)))))..)).....))))))))).)...... ( -32.00, z-score = 0.02, R) >dp4.chrXL_group1e 4608118 106 - 12523060 -CGCGUGGACGUAGCACUCGAGUGGUAAUCCUUAGACAUGUUUUUAGCUGCAAUCUAGCUCAAUCUAUCUGCGUAGACAUCUACAUAGGUGGGGCUACGUGGGCUGC---------- -.((((..(((((((....((((((....)).((((..(((........))).))))))))..((((((((.((((....)))).)))))))))))))))..)).))---------- ( -34.00, z-score = -1.13, R) >droPer1.super_13 652062 109 + 2293547 -CUCGUGGACGUAGCACUCGAGUGGUAAUCCUUAGACAUGUUUUUAGUUGCAAUCUAGCUCAAUCUAUCUGCGUAGACAUCUACAUAGGUGGGGCUACGUGGGCUGCUGC------- -...((..(((((((....((((((....)).((((..(((........))).))))))))..((((((((.((((....)))).)))))))))))))))..))......------- ( -33.20, z-score = -0.91, R) >droGri2.scaffold_15245 17767471 96 - 18325388 ---------------------UGUGUAGAACAUACGACUGUCUGUAUGCUCAACUCAGCCAGAUCUAACUGACUACAAAUAUAUAUAUAUACAUGUAUCUUUAUAUUUGUUAAACUG ---------------------.(.((.((.((((((......)))))).)).)).)((.(((......))).))(((((((((.(((((....)))))...)))))))))....... ( -16.50, z-score = -0.78, R) >consensus _UUCGGGUCUGUAGCUCCAAACGAGCGA_ACUCUUACUUGUUUC_AGCUGCAAUCUAGUCCAAUCUAUCUGCGUAGACAUCUGAAAAGGUGGGGCUACGUGGGCUGAUGCCAUUCUG ......................((((............(((........)))................((((((((.(((((....)))))...))))))))))))........... (-14.57 = -15.82 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:28 2011