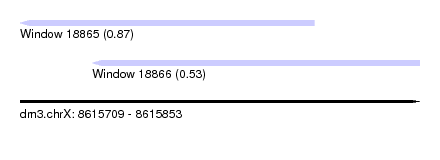
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,615,709 – 8,615,853 |
| Length | 144 |
| Max. P | 0.865425 |

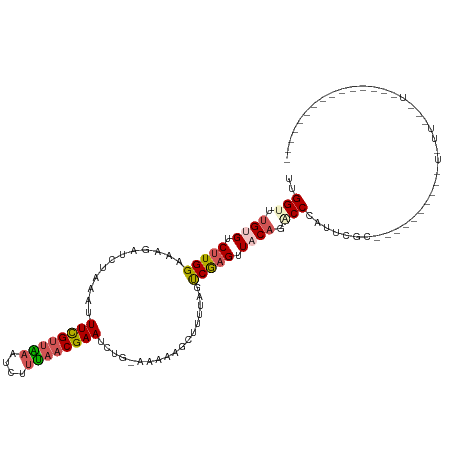
| Location | 8,615,709 – 8,615,815 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.04 |
| Shannon entropy | 0.64520 |
| G+C content | 0.35572 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.88 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8615709 106 - 22422827 UUGGUUUGUGCCCUGGAAAGAUCUAAAUUUCGUUAAAUAUUUAACGAAUCUGAAAAAAGCGUUUAGUCGAGUUACAUACCCAUUCGCUUUUACUUUUUCUUGUUUU-------------- ..(((....)))..(((((((((.....((((((((....))))))))...)).(((((((.......................)))))))..)))))))......-------------- ( -16.70, z-score = -0.14, R) >droEre2.scaffold_4690 12265594 118 + 18748788 UCGGUUUGUGUCUUGGCGAGAUCUAAGUUUCGUGGAACCACCGUCGAAUUU--UAAACGUUUUUAGUCGAGUUUCAGCCCCAUUCGCAAAAUUAAACUCUUGUUUAAUUUUUUUUUUAGU ..((.(((...(((((((((((.((((.(((((((.....))).)))).))--))...)))))..))))))...))).)).......((((((((((....))))))))))......... ( -23.40, z-score = -0.76, R) >droYak2.chrX 9074318 115 + 21770863 CCGGUUUGUGUCUUGGCAUGACCUAAAUUUCGUUGAAUCUUUAACGAAACU--UAAAACAUUUUAGUCGAGUUUCAACCCCAGUCGCAUAAUGGAACUUUUUACUUGUUUGUUUGAU--- ..((.(..(((....)))..)))(((.(((((((((....))))))))).)--))(((((..(.(((.((((((((...............))))))))...))).)..)))))...--- ( -23.26, z-score = -0.97, R) >droSec1.super_35 457627 86 - 471130 UUGGUAUGUGUCUUGGAAAGAUCUAAAUUUUGUUAAAUGUUUAACGAAUCUGAAAAAAGCUCUUAGCCGAGUUACAGGCCCAUACG---------------------------------- ..(((.((((.(((((.((((.((....((((((((....)))))))).........)).))))..))))).)))).)))......---------------------------------- ( -18.82, z-score = -1.74, R) >droSim1.chrX 6866896 85 - 17042790 UUGGUCAGUGUCUUGGAAAGAUCUAAAUUUCGUUAAAUGUUUAACGAAUCUG-AAAAAGCUCUUAGCCAAGUUACACACCCGUUCG---------------------------------- ..(((..(((((((((.((((.((....((((((((....))))))))....-....)).))))..)))))..)))))))......---------------------------------- ( -18.90, z-score = -2.08, R) >consensus UUGGUUUGUGUCUUGGAAAGAUCUAAAUUUCGUUAAAUCUUUAACGAAUCUG_AAAAAGCUUUUAGUCGAGUUACAGACCCAUUCGC__________U_UU___U_______________ ..(((.((((.(((((......(((((.((((((((....)))))))).............)))))))))).)))).)))........................................ ( -9.80 = -10.88 + 1.08)


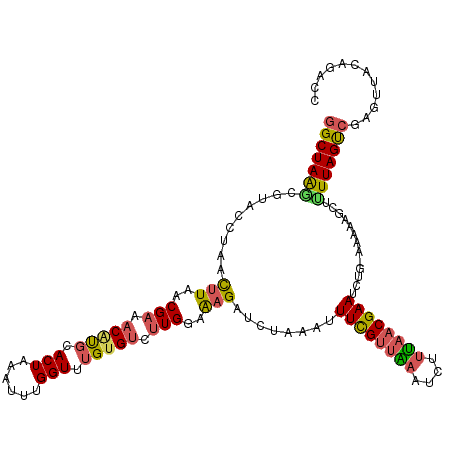
| Location | 8,615,735 – 8,615,853 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.33603 |
| G+C content | 0.38137 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8615735 118 - 22422827 GGCUAAGCGUACCUAACUUAACGAAACUCGCACUAAAUUUGGUUUGUGCCCUGGAAAGAUCUAAAUUUCGUUAAAUAUUUAACGAAUCUGAAAAAAGCGUUUAGUCGAGUUACAUACC ((((((((((.......(((((((((...((((............)))).((....)).......)))))))))...((((.......))))....))))))))))............ ( -25.20, z-score = -1.59, R) >droEre2.scaffold_4690 12265634 116 + 18748788 GGCUAAACGUUCCUAAUUUAACGAAACGUGCACUAAAAUCGGUUUGUGUCUUGGCGAGAUCUAAGUUUCGUGGAACCACCGUCGAAUUU--UAAACGUUUUUAGUCGAGUUUCAGCCC ((((....(((........)))(((((.((.(((((((...(((((....((((((((((....)))))(((....))).)))))....--)))))..))))))))).))))))))). ( -29.40, z-score = -1.20, R) >droYak2.chrX 9074355 116 + 21770863 CGCUAGGCGUUCCUAACUUAACGAAACGUGCACUAAAACCGGUUUGUGUCUUGGCAUGACCUAAAUUUCGUUGAAUCUUUAACGAAACU--UAAAACAUUUUAGUCGAGUUUCAACCC ...((((....)))).......(((((.((.(((((((..((.(..(((....)))..)))(((.(((((((((....))))))))).)--)).....))))))))).)))))..... ( -27.30, z-score = -2.02, R) >droSec1.super_35 457633 118 - 471130 GGCUAAACGUACCUAACUUAACGAAACAUGCACUAAAUUUGGUAUGUGUCUUGGAAAGAUCUAAAUUUUGUUAAAUGUUUAACGAAUCUGAAAAAAGCUCUUAGCCGAGUUACAGGCC ((((((..((......(((..(((.((((..((((....))))..)))).)))..))).((.....((((((((....))))))))...)).....))..))))))............ ( -23.20, z-score = -0.95, R) >droSim1.chrX 6866902 117 - 17042790 GGCUAAGCGUACCUAACUUAACGAAACAUGCACUAAAUUUGGUCAGUGUCUUGGAAAGAUCUAAAUUUCGUUAAAUGUUUAACGAAUCUG-AAAAAGCUCUUAGCCAAGUUACACACC (((((((.((......(((..(((.((((..((((....))))..)))).)))..))).((.....((((((((....))))))))...)-)....)).)))))))............ ( -27.80, z-score = -2.70, R) >consensus GGCUAAGCGUACCUAACUUAACGAAACAUGCACUAAAUUUGGUUUGUGUCUUGGAAAGAUCUAAAUUUCGUUAAAUCUUUAACGAAUCUG_AAAAAGCUUUUAGUCGAGUUACAGACC (((((((.........(((..(((.(((((.(((......))).))))).)))..)))........((((((((....)))))))).............)))))))............ (-16.12 = -16.28 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:24 2011