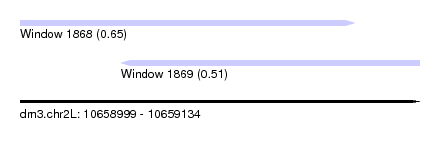
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,658,999 – 10,659,134 |
| Length | 135 |
| Max. P | 0.653031 |

| Location | 10,658,999 – 10,659,112 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Shannon entropy | 0.59516 |
| G+C content | 0.55114 |
| Mean single sequence MFE | -37.31 |
| Consensus MFE | -14.19 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
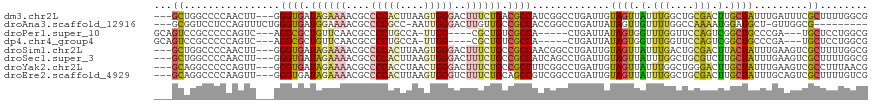
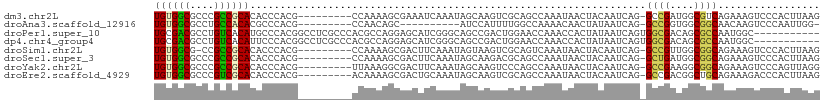
>dm3.chr2L 10658999 113 + 23011544 ---GCUGGCCCCAACUU---GGGUGAUAGAAAACGCCCCACUUAAGUGGGACUUUCUGACGCCAUCGGCCUGAUUGUAGUUAUUUGGCUGCGACUUGCUAUUUGAUUUCGCUUUUGGCG ---(((((((.......---.((((.((((((..(.(((((....))))).))))))).))))...)))).....(.(((((..((((........))))..))))).)......))). ( -34.10, z-score = -1.51, R) >droAna3.scaffold_12916 14502818 105 + 16180835 ---GCGGUCCUCCAGUUUCUGGGUGAUGGAAAACGCCCCGCC-AAUUGGGACUUGUUGCCGCCACCGGCCUGAUUAUAGUUGUUUUGGCCAAAAUGGAUGCU-GUUGGCG--------- ---...(.((..((((.((((((((.(....).))))).(((-((..(.((((.(((((((....))))..)))...)))).).)))))......))).)))-)..))).--------- ( -32.30, z-score = 0.40, R) >droPer1.super_10 1869993 103 + 3432795 GCAGUCCGCCCCCAGUC---AGGCGCUGUUCAACGCCCCUGCCA-UUGG----CGCUGUCGCCA-----CUGAUUAUAGUGGUUUGGUUCCAGUCGGCUGCCCGA---UGCUCCUGGCG ((((..((((.......---.))))))))....((((...((.(-((((----.((.(((((((-----(((....)))))))(((....)))..))).))))))---)))....)))) ( -35.90, z-score = -0.43, R) >dp4.chr4_group4 2857820 103 + 6586962 GCAGUCCGCCCCCAGUC---AGGCGCUGUUCAACGCCCCUGCCA-UUGG----CGCUGUCGCCA-----CUGAUUAUAGUGGUUUGGUUCCAGUCGGCUGCCCGA---UGCUCCUGGCG ((((..((((.......---.))))))))....((((...((.(-((((----.((.(((((((-----(((....)))))))(((....)))..))).))))))---)))....)))) ( -35.90, z-score = -0.43, R) >droSim1.chr2L 10455839 113 + 22036055 ---GCUGGCCCCAACUU---GGGUGAUAGAAAACGCCCCACUUAAGUGGGACUUUCUGCCGCCAACGGCCUGAUUGUAGUUAUUUGACUGCGACUUACUAUUUGAAGUCGCUUUUGGCG ---...((((((.....---)))....(((((..(.(((((....))))).)))))))))(((((.(((......((((((....))))))(((((........)))))))).))))). ( -40.60, z-score = -3.87, R) >droSec1.super_3 6076842 113 + 7220098 ---GCUGGCCCCAACUU---GGGUGAUAGAAAACGCCCCACUUAAGUGGGACUUUCUGCCGCCAUCAGCCUGAUUGUAGUUAUUUGGCUGCGUCUUGCUAUUUGAAGUCGCUUUUGGCG ---...((((((.....---)))....(((((..(.(((((....))))).)))))))))((((..((((.(((.......))).))))(((.(((........))).)))...)))). ( -36.10, z-score = -1.38, R) >droYak2.chr2L 7062689 113 + 22324452 ---GCAGGCCCCCAGUU---GGGUGAUAGAAAACGCCCCACCUAACUGGGACUUUCUGCCGCCUUCGGCCUGAUUGUAGUUAUUUGGCUGGGACUUGCUAUUUGAAGUCGCCUUUAACG ---.(((((((((((((---(((((.............)))))))))))).(.....)........))))))......((((...(((...(((((........))))))))..)))). ( -40.92, z-score = -2.61, R) >droEre2.scaffold_4929 11867346 113 - 26641161 ---GCAGGCCCCAAGUU---GGGUGAUAGAAAACGCCCCACUUAAGUGGGUCUUUCUGCAGCCGUCGGCCUGAUUGUAGUUAUUUGGCUGCGACUUGCUAUUUGCAGUCGCUUUUGUCG ---.((((((......(---((.((.((((((..(.(((((....))))).))))))))).)))..)))))).............(((.((((((.((.....))))))))....))). ( -42.70, z-score = -2.76, R) >consensus ___GCUGGCCCCAAGUU___GGGUGAUAGAAAACGCCCCACCUAAGUGGGACUUUCUGCCGCCAUCGGCCUGAUUGUAGUUAUUUGGCUGCGACUUGCUAUUUGAAGUCGCUUUUGGCG ...((................((((.((((((....(((((....)))))..)))))).)))).............((((.(.(((....))).).)))).........))........ (-14.19 = -13.78 + -0.42)
| Location | 10,659,033 – 10,659,134 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.79 |
| Shannon entropy | 0.62973 |
| G+C content | 0.56811 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -12.20 |
| Energy contribution | -11.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
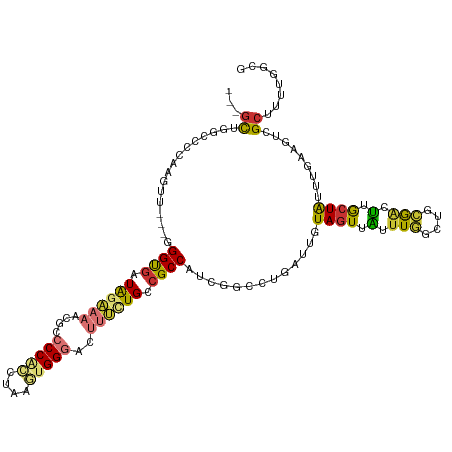
>dm3.chr2L 10659033 101 - 23011544 UGUGGCGCCCGCCGCACACCCACG---------CCAAAAGCGAAAUCAAAUAGCAAGUCGCAGCCAAAUAACUACAAUCAG-GCCGAUGGCGUCAGAAAGUCCCACUUAAG ((((((....)))))).....(((---------(((...((((...(.....)....)))).(((...............)-))...)))))).................. ( -24.36, z-score = -1.44, R) >droAna3.scaffold_12916 14502855 90 - 16180835 UGUGGCGCCUGCCACACGCCCACG---------CCAACAGC----------AUCCAUUUUGGCCAAAACAACUAUAAUCAG-GCCGGUGGCGGCAACAAGUCCCAAUUGG- ((((((....)))))).(((..((---------((....(.----------...).....((((................)-)))))))..)))................- ( -24.39, z-score = 0.54, R) >droPer1.super_10 1870030 100 - 3432795 UGCGACGCCUGUCACAUGCCCACGGCCUCGCCCACGCCAGGAGCAUCGGGCAGCCGACUGGAACCAAACCACUAUAAUCAGUGGCGACAGCGCCAAUGGC----------- ......(((((((......(((((((...((((..((.....))...)))).))))..))).......(((((......))))).))))).)).......----------- ( -32.10, z-score = 0.06, R) >dp4.chr4_group4 2857857 100 - 6586962 UGCGACGCCUGUCACAUUCCCACGGCCUCGCCCACGCCAGGAGCAUCGGGCAGCCGACUGGAACCAAACCACUAUAAUCAGUGGCGACAGCGCCAAUGGC----------- ......(((((((...((((..((((...((((..((.....))...)))).))))...)))).....(((((......))))).))))).)).......----------- ( -33.20, z-score = -0.79, R) >droSim1.chr2L 10455873 100 - 22036055 UGUGGCG-CCGCCGCACACCCACG---------CCAAAAGCGACUUCAAAUAGUAAGUCGCAGUCAAAUAACUACAAUCAG-GCCGUUGGCGGCAGAAAGUCCCACUUAAG .((((.(-(.(((((.((...(((---------((....((((((((.....).)))))))(((......))).......)-).)))))))))).....)).))))..... ( -30.30, z-score = -2.62, R) >droSec1.super_3 6076876 101 - 7220098 UGUGGCGCCCGCCGCACACCCACG---------CCAAAAGCGACUUCAAAUAGCAAGACGCAGCCAAAUAACUACAAUCAG-GCUGAUGGCGGCAGAAAGUCCCACUUAAG ((((((....))))))......((---------(((...(((.(((........))).)))((((...............)-)))..)))))................... ( -26.36, z-score = -1.01, R) >droYak2.chr2L 7062723 101 - 22324452 UGUGGCGCCCGCCGCACACCCACG---------UUAAAGGCGACUUCAAAUAGCAAGUCCCAGCCAAAUAACUACAAUCAG-GCCGAAGGCGGCAGAAAGUCCCAGUUAGG ((((((....))))))...((...---------.....((((((((........)))))...)))...(((((((..((..-((((....)))).))..))...))))))) ( -26.20, z-score = -0.60, R) >droEre2.scaffold_4929 11867380 101 + 26641161 UGUGGCGCCCGUCGCACACCCACG---------ACAAAAGCGACUGCAAAUAGCAAGUCGCAGCCAAAUAACUACAAUCAG-GCCGACGGCUGCAGAAAGACCCACUUAAG ....(((.((((((..........---------......((((((((.....)).)))))).(((...............)-)))))))).)))................. ( -28.46, z-score = -2.17, R) >consensus UGUGGCGCCCGCCGCACACCCACG_________CCAAAAGCGACUUCAAAUAGCAAGUCGCAGCCAAAUAACUACAAUCAG_GCCGAUGGCGGCAGAAAGUCCCACUUAAG ((((((....))))))..................................................................((((....))))................. (-12.20 = -11.80 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:40 2011