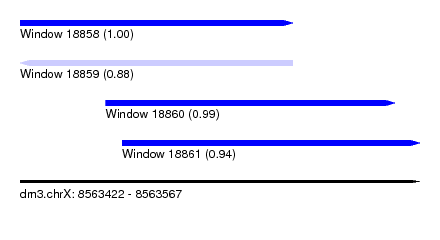
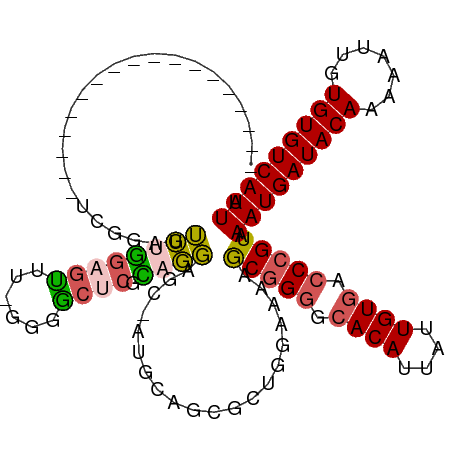
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,563,422 – 8,563,567 |
| Length | 145 |
| Max. P | 0.998058 |

| Location | 8,563,422 – 8,563,521 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.51149 |
| G+C content | 0.45685 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8563422 99 + 22422827 ------------------UUGGAGUUUGGAGUUUUGGGACUCGCAGGGGC-AUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA ------------------...((((((.((...)).))))))(((.....-.)))..((((.....))))(((((((....)))).)))...(((((((((.......))))))))). ( -32.00, z-score = -2.45, R) >droSim1.chrX 6810873 98 + 17042790 ------------------UCGGAGUUUGGAGUUU-GGGGCUCGCAGGAGA-AUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA ------------------.............((.-.(.(((.(((.....-.)))))).)..))...(((((.((((....)))).))))).(((((((((.......))))))))). ( -33.60, z-score = -3.07, R) >droSec1.super_35 409939 98 + 471130 ------------------UCGGAGUUUGAAGUUU-GGGGCUCGCAGGAGA-AUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA ------------------.............((.-.(.(((.(((.....-.)))))).)..))...(((((.((((....)))).))))).(((((((((.......))))))))). ( -33.60, z-score = -3.18, R) >droAna3.scaffold_13248 801180 107 - 4840945 -----------UCGGGAGGUGGAGAUCUGAUUUCGGAUGCCCAAAGUCCCCAAGAGGCGAUCGAAGAACGGGCCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA -----------..((((..(((..(((((....)))))..)))...)))).................(((((.((((....)))).))))).(((((((((.......))))))))). ( -36.50, z-score = -2.83, R) >droWil1.scaffold_181150 3925043 118 + 4952429 UCGGAGUCAGCUUGGUCAGCAGCUUAAAGAGCCAGAAUGCGCUUCUGUGCAACGCCACGAAAAGUUAACCGAAAACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA .(((.(((...(((((.(((.((((...)))).....(((((....)))))............))).)))))..(((....)))))))))..(((((((((.......))))))))). ( -31.30, z-score = -1.79, R) >consensus __________________UCGGAGUUUGGAGUUU_GGGGCUCGCAGGAGC_AUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUA ........................((((((((......)))).))))....................(((((.((((....)))).))))).(((((((((.......))))))))). (-19.68 = -19.72 + 0.04)

| Location | 8,563,422 – 8,563,521 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.51149 |
| G+C content | 0.45685 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.36 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8563422 99 - 22422827 UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUGCCCCGCUUUUCCAGCGCUGCAU-GCCCCUGCGAGUCCCAAAACUCCAAACUCCAA------------------ .(((((.(((.......))).)))))...(((.((((....)))))))((((.....))))..(((.-.....)))((((......))))..........------------------ ( -19.40, z-score = -1.54, R) >droSim1.chrX 6810873 98 - 17042790 UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUGCCCCGCUUUUCCAGCGCUGCAU-UCUCCUGCGAGCCCC-AAACUCCAAACUCCGA------------------ .(((((.(((.......))).)))))...(((.((((....)))).)))(((.....)))((((((.-.....))).)))...-................------------------ ( -19.80, z-score = -1.81, R) >droSec1.super_35 409939 98 - 471130 UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUGCCCCGCUUUUCCAGCGCUGCAU-UCUCCUGCGAGCCCC-AAACUUCAAACUCCGA------------------ .(((((.(((.......))).)))))...(((.((((....)))).)))(((.....)))((((((.-.....))).)))...-................------------------ ( -19.80, z-score = -1.72, R) >droAna3.scaffold_13248 801180 107 + 4840945 UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUGGCCCGUUCUUCGAUCGCCUCUUGGGGACUUUGGGCAUCCGAAAUCAGAUCUCCACCUCCCGA----------- .(((((.(((.......))).))))).((((((((((....)))))))))).....((((((((...(....)...))))....(....).))))............----------- ( -31.10, z-score = -2.02, R) >droWil1.scaffold_181150 3925043 118 - 4952429 UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUUUUCGGUUAACUUUUCGUGGCGUUGCACAGAAGCGCAUUCUGGCUCUUUAAGCUGCUGACCAAGCUGACUCCGA .(((((.(((.......))).)))))..((((((.....((((((..(((........))).))))))((.(((((.....)))))))......((((........))))))).))). ( -26.80, z-score = -0.31, R) >consensus UAAUGACACACAAUUUUUGUAUCAUUUACGGGUCACAAUAAUGUGCCCCGCUUUUCCAGCGCUGCAU_GCUCCUGCGAGCCCC_AAACUCCAAACUCCGA__________________ .(((((.(((.......))).)))))..((((.((((....)))).))))..........(((..............)))...................................... (-13.00 = -12.36 + -0.64)

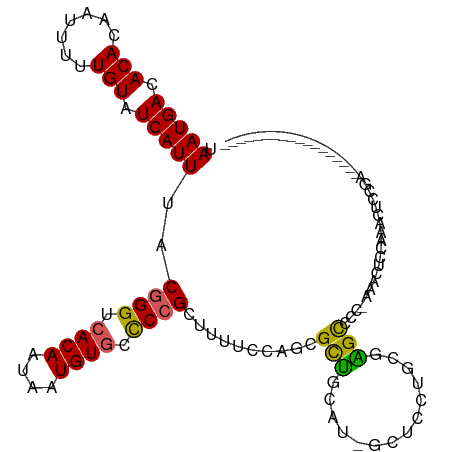
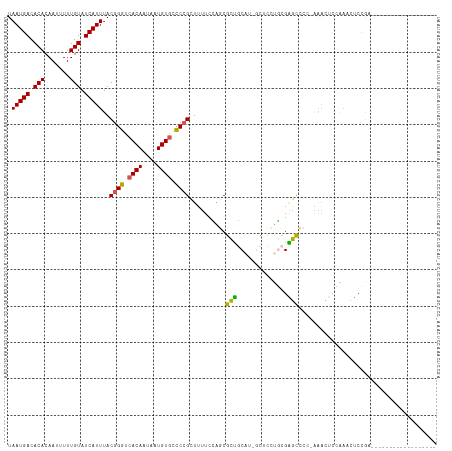
| Location | 8,563,453 – 8,563,558 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.67 |
| Shannon entropy | 0.58242 |
| G+C content | 0.46789 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8563453 105 + 22422827 ------------CAUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGAAGGGGCGG-- ------------(((((.(.((((.....(((((.((((....)))).))))).(((((((((.......)))))))))......)))).).)))))((.(((.......)))))..-- ( -39.50, z-score = -3.81, R) >droSim1.chrX 6810903 104 + 17042790 ------------AAUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGA-GGGGCGG-- ------------.((((.(.((((.....(((((.((((....)))).))))).(((((((((.......)))))))))......)))).).)))).((.(((......-)))))..-- ( -37.10, z-score = -3.12, R) >droSec1.super_35 409969 104 + 471130 ------------AAUGCAGCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGA-GGGGCGG-- ------------.((((.(.((((.....(((((.((((....)))).))))).(((((((((.......)))))))))......)))).).)))).((.(((......-)))))..-- ( -37.10, z-score = -3.12, R) >droYak2.chrX 9022525 104 - 21770863 ------------AACGUGGCGAUCGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCGUGGCAUCCACCAGGGGGGGGC--- ------------...(((.((........(((((.((((....)))).))))).(((((((((.......)))))))))......)).)))......((..((.(....)))..))--- ( -33.50, z-score = -1.82, R) >droEre2.scaffold_4690 12197893 101 - 18748788 ------------AACGCAGCAAUCGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAGUUGUGUGUCAUUAAUAAACGGCACAGCGUGGCAUCCU-CAGG-GGGGC---- ------------.((((.((.........(((((.((((....)))).))))).(((((((((.......)))))))))........))...)))).((..((.-...)-)..))---- ( -31.40, z-score = -2.03, R) >droAna3.scaffold_13248 801216 108 - 4840945 ---------CCCCAAGAGGCGAUCGAAGAACGGGCCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCAGCACAGCGUGGCAUUUAUGAGGAGGGAAAG-- ---------.(((..(..((.........(((((.((((....)))).))))).(((((((((.......))))))))).....))..).(..((((.....))))..).)))....-- ( -30.40, z-score = -2.44, R) >dp4.chrXL_group1e 7800830 86 + 12523060 -----------------------------UGGGGCAACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCAUGG----CCUCCACAGAGAGCCAG -----------------------------..(((..(((....)))..)))...(((((((((.......))))))))).....((......)).(((----((((....))).)))). ( -26.30, z-score = -2.26, R) >droPer1.super_13 1541034 86 - 2293547 -----------------------------UGGGGCAACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCAUGG----CCUCCACAGAGAGCGAG -----------------------------((((((..(((..(((((..((((.(((((((((.......))))))))).....))))))))).))))----)).)))........... ( -22.60, z-score = -1.19, R) >droWil1.scaffold_181150 3925081 119 + 4952429 GCGCUUCUGUGCAACGCCACGAAAAGUUAACCGAAAACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCAUGGUAUCCCGCCACAGAGAGAGAG .(.(((((((((..((((((((.(((((.......))).)).))))).......(((((((((.......))))))))).....)))))))))..((((.....))))..))).).... ( -26.50, z-score = -0.80, R) >droVir3.scaffold_12970 5324717 80 - 11907090 --------------------AGCUUU---GCAGCGAACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCGUGGCAUCC---------------- --------------------.(((.(---((.((..(((....)))........(((((((((.......))))))))).....)).))).))).........---------------- ( -19.90, z-score = -0.76, R) >droGri2.scaffold_14853 5914353 88 + 10151454 ------------AGGUCCGCAGCCGCA--GCAGAACAUAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCGUG-CAUCC---------------- ------------..(..(((.(((((.--......((((....)))).......(((((((((.......))))))))).....)))))...)))..-)....---------------- ( -26.20, z-score = -2.48, R) >consensus ____________AA_GCAGCGAUCGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCAUGGCAUCCCGCAGA_GGGGC_G__ .....................................(((..(((((..((((.(((((((((.......))))))))).....))))))))).)))...................... (-15.29 = -14.80 + -0.49)


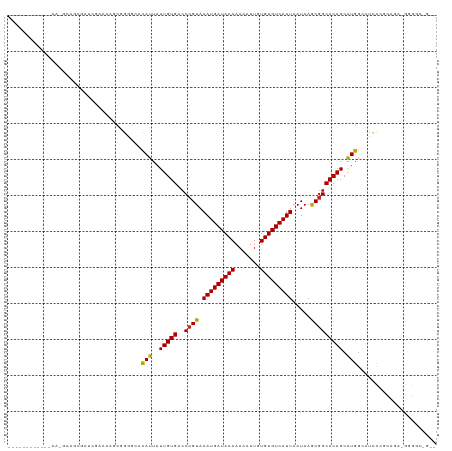
| Location | 8,563,459 – 8,563,567 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.38392 |
| G+C content | 0.49261 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8563459 108 + 22422827 GCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGAAGGGGCGGGGGGGUGAG ..((((.....(((((.((((....)))).))))).(((((((((.......)))))))))...........)))).(.((.((((.(......).))))...)).). ( -36.00, z-score = -2.65, R) >droSim1.chrX 6810909 104 + 17042790 GCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGAGGGGCGGGGGGAG---- ..((((.....(((((.((((....)))).))))).(((((((((.......)))))))))...........))))......(((.((......)).)))....---- ( -34.60, z-score = -2.68, R) >droSec1.super_35 409975 104 + 471130 GCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCAUGGCAUCCGGCAGAGGGGCGGGGGGAG---- ..((((.....(((((.((((....)))).))))).(((((((((.......)))))))))...........))))......(((.((......)).)))....---- ( -34.60, z-score = -2.68, R) >droYak2.chrX 9022531 104 - 21770863 GCGAUCGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCGUGGCAUCCACCAGGGGGGGGCGGGGAA---- ((.........(((((.((((....)))).))))).(((((((((.......)))))))))........))....(.(.((..((.(....)))..)).).)..---- ( -31.20, z-score = -1.12, R) >droEre2.scaffold_4690 12197899 101 - 18748788 GCAAUCGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAGUUGUGUGUCAUUAAUAAACGGCACAGCGUGGCAUCCU-CAGGGGGGCGGGGUA------ ((.........(((((.((((....)))).))))).(((((((((.......)))))))))........))...((.(.((..((.-...))..)).).)).------ ( -32.30, z-score = -1.98, R) >droAna3.scaffold_13248 801225 105 - 4840945 GCGAUCGAAGAACGGGCCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCAGCACAGCGUGGCAUUUAUGAGGAGGGAAAGGGGCUA--- ((..((.....(((((.((((....)))).))))).(((((((((.......))))))))).....((.((.....)).)).................)).))..--- ( -25.30, z-score = -0.58, R) >droGri2.scaffold_14853 5914359 82 + 10151454 --GCAGCCGCAGCAGAACAUAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAGCGGCACAGCGUG-CAUCC----------------------- --((.(((((.......((((....)))).......(((((((((.......))))))))).....)))))...))...-.....----------------------- ( -21.90, z-score = -1.76, R) >consensus GCGCUGGAAAAGCGGGGCACAUUAUUGUGACCCGUAAAUGAUACAAAAAUUGUGUGUCAUUAAUAAACGGCACAGCGUGGCAUCCGGCAGAGGGGCGGGGGGAG____ ((.........(((((.((((....)))).))))).(((((((((.......))))))))).............))................................ (-20.83 = -21.16 + 0.33)

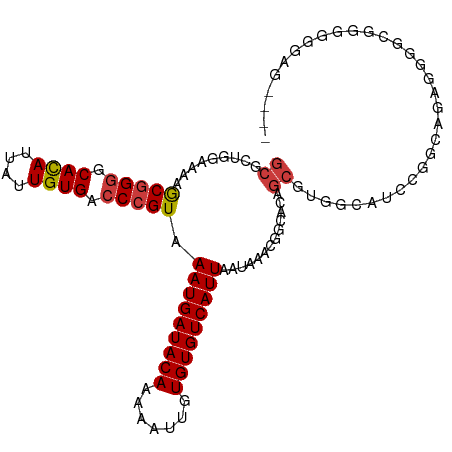
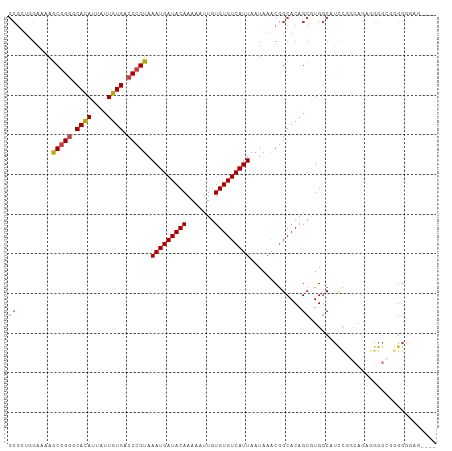
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:19 2011