| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,550,331 – 8,550,461 |
| Length | 130 |
| Max. P | 0.636608 |

| Location | 8,550,331 – 8,550,461 |
|---|---|
| Length | 130 |
| Sequences | 10 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.53406 |
| G+C content | 0.50439 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
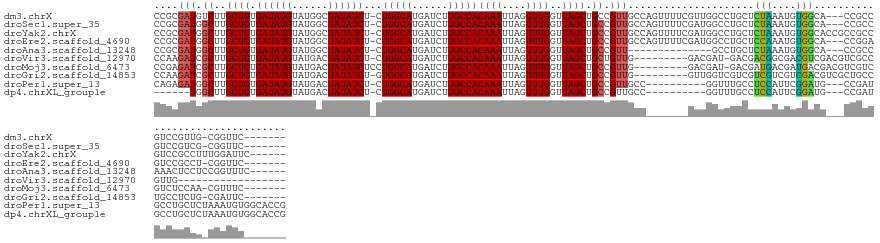
>dm3.chrX 8550331 130 + 22422827 -------GAACCG-CAACGGACGGCGG---UGCCACAUUUAGAGCAGGCCAACGAAAACUGGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGCCAUACAUAUCACAGCAAGACAUCGCGG -------...(((-(.......((((.---(((((((.((((.....((((........))))...((.......))...))))..)))))))......))))..-.(((((........))))).............)))) ( -35.40, z-score = -1.92, R) >droSec1.super_35 397832 130 + 471130 -------GAACCG-CGACGGACGGCGG---UGCCACAUUUAGAGCAGGCCAUCGAAAACUGGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGCCAUACAUAUCACAGCAAGCCAUCGCGG -------...(((-(((.((.(((((.---(((((((.((((.....((((........))))...((.......))...))))..)))))))......))))..-.(((((........))))).......))).)))))) ( -41.70, z-score = -2.87, R) >droYak2.chrX 9010222 135 - 21770863 ------GAAUCCAAAGGCGGACGGCGGCGGUGCCACAUUUAGAGCAGGCCAUCGAAAACUGGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGCCAUACAUAUCACAGCAAGCCAUCGCGG ------..........((((..(((((((.(((((((.((((.....((((........))))...((.......))...))))..)))))))......))))..-.(((((........))))).......))).)))).. ( -38.20, z-score = -0.97, R) >droEre2.scaffold_4690 12185700 130 - 18748788 -------GAACCG-AGGCGGACUCCGG---UGCCACAUUUGGAGCAGGCCAUCGAAAACUGGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGCCAUACAUAUCACAGCAAGCCAUCGCGG -------...(((-((((((.(((((.---(((((..(((.((........)).)))..))))).))).)))).......(((((((.(((((......))))).-)))).)))..................))).))).). ( -35.00, z-score = -0.45, R) >droAna3.scaffold_13248 782540 118 - 4840945 ------GAAACCGGAGGAGUUUGGCGG---UGCCACAUUUAGAGCAGGC--------------AACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGCCAUACAUAUCACAGCAAGCCAUCGCGG ------....(((((((..(((((((.---(((((((.((((....(..--------------..)((.......))...))))..)))))))......))))))-)(((((........)))))........)).)).))) ( -33.10, z-score = -1.54, R) >droVir3.scaffold_12970 6594404 113 + 11907090 ------------------CAACGGCGACGUCGACGUCGCCGUCGUC-AUCGUC---------CAACAGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGUCAUACAUAUCACAGCAAGCGAUCUUGG ------------------..(((((((((....)))))))))....-.....(---------(((..((.(((.............(.(((((......))))).-)(((((........)))))..)))..))....)))) ( -32.30, z-score = -2.34, R) >droMoj3.scaffold_6473 15571915 124 - 16943266 -------GAAACG-UUGGAGACGACGACGUCGUCAUCGUCAUCGUC-AUCGUC---------CAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAGGAGAUAUAGUCAUACAUAUCACAGCAAGCGAUCUCGG -------....((-(((((((.(((((.(.((....)).).)))))-.)).))---------)))))((.(((............((.(((((......))))).))(((((........)))))..)))..))........ ( -36.60, z-score = -2.29, R) >droGri2.scaffold_14853 4248735 124 - 10151454 -------GAAUCG-CAGAGGCAGGCAGCGACGUCGACGACGACGACGACCAAC---------CAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAC-AGAUAUAGUCAUACAUAUCACAGCAAGCGAUCUUGG -------..((((-(...((..(((..((.(((((....))))).)).((...---------....))..)))..))..((((((((((((((......))))))-)))).)))).................)))))..... ( -35.90, z-score = -3.12, R) >droPer1.super_13 1528891 128 - 2293547 CGGUGCCACAUUUAGAGCAGGCAUCGG---CAUCCGAAUGGAGGCAAACC----------GGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGUCAUACAUAUCACAGCAAGCCAUCUCUG .((((((............))))))((---(.......(((.(((...((----------(....)))..)))..))).((((((((.(((((......))))).-)))).)))).................)))....... ( -37.30, z-score = -1.93, R) >dp4.chrXL_group1e 7788891 122 + 12523060 CGGUGCCACAUUUAGAGCAGGCAUCGG---CAUCCGAAUGGAGGCAAACC----------GGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG-AGAUAUAGUCAUACAUAUCACAGCAAGCCA------ .((((((............))))))((---(.......(((.(((...((----------(....)))..)))..))).((((((((.(((((......))))).-)))).)))).................))).------ ( -37.30, z-score = -2.60, R) >consensus _______GAACCG_AAGCGGACGGCGG___CGCCACAUUUAGAGCAGACCAUC_______GGCAACGGCAGCUAACCAAACUAAUUUGUGGCAAGAUCAUGCCAG_AGAUAUAGCCAUACAUAUCACAGCAAGCCAUCGCGG ......................................................................(((...............(((((......)))))...(((((........)))))..)))............ (-12.07 = -12.07 + 0.00)



| Location | 8,550,331 – 8,550,461 |
|---|---|
| Length | 130 |
| Sequences | 10 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.53406 |
| G+C content | 0.50439 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -16.54 |
| Energy contribution | -15.34 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
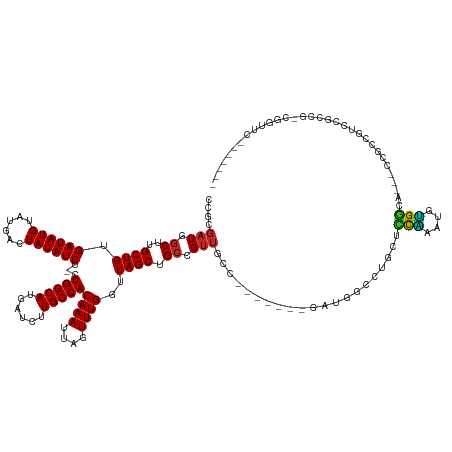
>dm3.chrX 8550331 130 - 22422827 CCGCGAUGUCUUGCUGUGAUAUGUAUGGCUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCCAGUUUUCGUUGGCCUGCUCUAAAUGUGGCA---CCGCCGUCCGUUG-CGGUUC------- ((((((((((.(((((.((((((......)))))).-).)))).))...(((((((..((((...(((.....)))((.(((((......)))))..)).)))).)))))))---........)))))-)))...------- ( -41.30, z-score = -1.56, R) >droSec1.super_35 397832 130 - 471130 CCGCGAUGGCUUGCUGUGAUAUGUAUGGCUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCCAGUUUUCGAUGGCCUGCUCUAAAUGUGGCA---CCGCCGUCCGUCG-CGGUUC------- ((((((((((.(((((.((((((......)))))).-).))))......(((((((......(((((..(((.(((((((........)))))))..)))))))))))))))---.....).))))))-)))...------- ( -47.60, z-score = -2.50, R) >droYak2.chrX 9010222 135 + 21770863 CCGCGAUGGCUUGCUGUGAUAUGUAUGGCUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCCAGUUUUCGAUGGCCUGCUCUAAAUGUGGCACCGCCGCCGUCCGCCUUUGGAUUC------ ..((((((((.(((((.((((((......)))))).-).))))......(((((((......(((((..(((.(((((((........)))))))..))))))))))))))).....))))).)))..........------ ( -42.40, z-score = -0.84, R) >droEre2.scaffold_4690 12185700 130 + 18748788 CCGCGAUGGCUUGCUGUGAUAUGUAUGGCUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCCAGUUUUCGAUGGCCUGCUCCAAAUGUGGCA---CCGGAGUCCGCCU-CGGUUC------- ...(((.(((..((((((.....)))))).....((-((((........(((((((......(((((..(((.(((((((........)))))))..)))))))))))))))---))))))...))))-))....------- ( -43.50, z-score = -1.16, R) >droAna3.scaffold_13248 782540 118 + 4840945 CCGCGAUGGCUUGCUGUGAUAUGUAUGGCUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUU--------------GCCUGCUCUAAAUGUGGCA---CCGCCAAACUCCUCCGGUUUC------ ((((((((((..((((.((((((......)))))).-.(((((......)))))((((....))))..)))).))))))--------------))............((((.---..)))).........))....------ ( -32.60, z-score = -0.58, R) >droVir3.scaffold_12970 6594404 113 - 11907090 CCAAGAUCGCUUGCUGUGAUAUGUAUGACUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCUGUUG---------GACGAU-GACGACGGCGACGUCGACGUCGCCGUUG------------------ .((((((((..(((((.((((((......)))))).-).))))))))))))(((((...(((((.....))))).)).))---------).....-..(((((((((((....)))))))))))------------------ ( -41.30, z-score = -3.24, R) >droMoj3.scaffold_6473 15571915 124 + 16943266 CCGAGAUCGCUUGCUGUGAUAUGUAUGACUAUAUCUCCUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUG---------GACGAU-GACGAUGACGAUGACGACGUCGUCGUCUCCAA-CGUUUC------- .((((((((..(((((.((.(((((....))))).)).)))))))))))))........(((((.....))))).(((((---------((.(((-(((((((.((....)).)))))))))))))))-))....------- ( -44.00, z-score = -3.49, R) >droGri2.scaffold_14853 4248735 124 + 10151454 CCAAGAUCGCUUGCUGUGAUAUGUAUGACUAUAUCU-GUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUG---------GUUGGUCGUCGUCGUCGUCGACGUCGCUGCCUGCCUCUG-CGAUUC------- ....((((((..((((.((((((......))))))(-((((((......)))))))...))))..(((.....)))...(---------((.(((((.(((((....))))).))..))).)))...)-))))).------- ( -41.60, z-score = -2.32, R) >droPer1.super_13 1528891 128 + 2293547 CAGAGAUGGCUUGCUGUGAUAUGUAUGACUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCC----------GGUUUGCCUCCAUUCGGAUG---CCGAUGCCUGCUCUAAAUGUGGCACCG ((((((((.(.(((........))).)....)))))-))).........(((((((..((((...(((.....)))...((.----------(((..((.(((....))).)---)....))).)).)))).)))))))... ( -37.00, z-score = -0.52, R) >dp4.chrXL_group1e 7788891 122 - 12523060 ------UGGCUUGCUGUGAUAUGUAUGACUAUAUCU-CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCC----------GGUUUGCCUCCAUUCGGAUG---CCGAUGCCUGCUCUAAAUGUGGCACCG ------.((..(((((.((((((......)))))).-).))))......(((((((..((((...(((.....)))...((.----------(((..((.(((....))).)---)....))).)).)))).))))))))). ( -34.40, z-score = -0.55, R) >consensus CCGCGAUGGCUUGCUGUGAUAUGUAUGACUAUAUCU_CUGGCAUGAUCUUGCCACAAAUUAGUUUGGUUAGCUGCCGUUGCC_______GAUGGCCUGCUCCAAAUGUGGCA___CCGCCGUCCGCCG_CGGUUC_______ ....(((.((..((((.((((((......))))))...(((((......)))))((((....))))..)))).)).))).............((......(((....))).....))......................... (-16.54 = -15.34 + -1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:13 2011