| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,637,988 – 10,638,049 |
| Length | 61 |
| Max. P | 0.964506 |

| Location | 10,637,988 – 10,638,049 |
|---|---|
| Length | 61 |
| Sequences | 6 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 68.04 |
| Shannon entropy | 0.62924 |
| G+C content | 0.39648 |
| Mean single sequence MFE | -12.08 |
| Consensus MFE | -6.39 |
| Energy contribution | -6.75 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964506 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
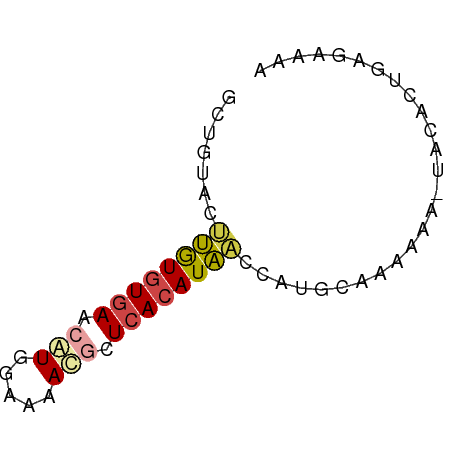
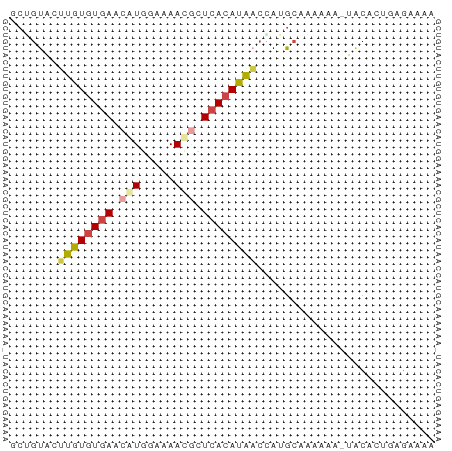
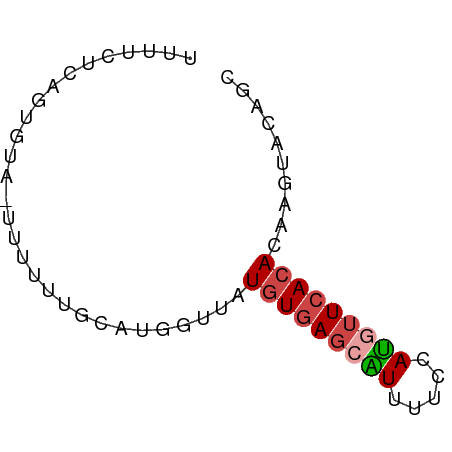
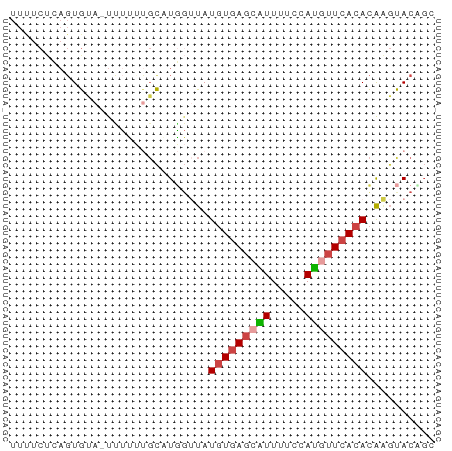
>dm3.chr2L 10637988 61 + 23011544 GCUGUACUUGUGUGAACAUGGAAAACGCUCACAUAACCAGGCAAAAAA-UACACUCAGAAAA (((....((((((((...((.....)).))))))))...)))......-............. ( -10.40, z-score = -1.04, R) >droSim1.chr2L 10435141 61 + 22036055 CCGGUACUUGUGUGAACAUGGAAAAUGCUCACAUAACCAUGCACAAAA-UACACUGAGAAAA ..(((...(((((((.(((.....))).))))))))))..........-............. ( -12.40, z-score = -1.75, R) >droSec1.super_3 6056119 61 + 7220098 GCGGUACUUGUGUGAACAUGGAAAAUGCUCACAUAACCAUGCAAAAAA-UACACUGAGAGAA (((....((((((((.(((.....))).))))))))...)))......-............. ( -13.50, z-score = -2.09, R) >droYak2.chr2L 7040839 62 + 22324452 GCUGUACUUGUGUGAACAUGGAAAACGCUCACAUAACCAUGCAAAAGAGUGCACUGAGAAAA ..(((((((...((..(((((...............)))))))...)))))))......... ( -12.16, z-score = -0.59, R) >droAna3.scaffold_12916 14479891 61 + 16180835 UGUGUGCCUGUGUGAAAAUGCAAAAUACUCACAUACGAAGACGGCUAA-UGCACUUGGAGAA .((((((((((((((..((.....))..))))))).......)))...-.))))........ ( -11.01, z-score = -0.20, R) >droVir3.scaffold_12963 9854807 51 - 20206255 GCAGUGGACAUGUAAGUGUUGAUAACUAUAA-AUGUCUAAGGCAAAAG-UUCA--------- ((..(((((((...(((.......)))....-)))))))..)).....-....--------- ( -13.00, z-score = -2.59, R) >consensus GCUGUACUUGUGUGAACAUGGAAAACGCUCACAUAACCAUGCAAAAAA_UACACUGAGAAAA .......((((((((.(((.....))).)))))))).......................... ( -6.39 = -6.75 + 0.36)

| Location | 10,637,988 – 10,638,049 |
|---|---|
| Length | 61 |
| Sequences | 6 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 68.04 |
| Shannon entropy | 0.62924 |
| G+C content | 0.39648 |
| Mean single sequence MFE | -11.85 |
| Consensus MFE | -6.45 |
| Energy contribution | -6.90 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950919 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
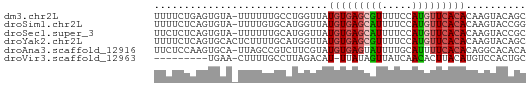
>dm3.chr2L 10637988 61 - 23011544 UUUUCUGAGUGUA-UUUUUUGCCUGGUUAUGUGAGCGUUUUCCAUGUUCACACAAGUACAGC ....(((((.(((-.....))))).....(((((((((.....)))))))))......))). ( -12.40, z-score = -1.00, R) >droSim1.chr2L 10435141 61 - 22036055 UUUUCUCAGUGUA-UUUUGUGCAUGGUUAUGUGAGCAUUUUCCAUGUUCACACAAGUACCGG ........(.(((-((((((((((((..(((....)))...)))))..)))).))))))).. ( -15.30, z-score = -2.15, R) >droSec1.super_3 6056119 61 - 7220098 UUCUCUCAGUGUA-UUUUUUGCAUGGUUAUGUGAGCAUUUUCCAUGUUCACACAAGUACCGC ........(((((-.....)))))(((..(((((((((.....))))))))).....))).. ( -15.40, z-score = -2.72, R) >droYak2.chr2L 7040839 62 - 22324452 UUUUCUCAGUGCACUCUUUUGCAUGGUUAUGUGAGCGUUUUCCAUGUUCACACAAGUACAGC ........(((((......))))).....(((((((((.....))))))))).......... ( -13.80, z-score = -1.21, R) >droAna3.scaffold_12916 14479891 61 - 16180835 UUCUCCAAGUGCA-UUAGCCGUCUUCGUAUGUGAGUAUUUUGCAUUUUCACACAGGCACACA ........((((.-.....((....))..((((((...........))))))...))))... ( -10.10, z-score = -0.47, R) >droVir3.scaffold_12963 9854807 51 + 20206255 ---------UGAA-CUUUUGCCUUAGACAU-UUAUAGUUAUCAACACUUACAUGUCCACUGC ---------....-.....((....(((((-.((.(((.......))))).)))))....)) ( -4.10, z-score = 0.07, R) >consensus UUUUCUCAGUGUA_UUUUUUGCAUGGUUAUGUGAGCAUUUUCCAUGUUCACACAAGUACAGC .............................(((((((((.....))))))))).......... ( -6.45 = -6.90 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:38 2011