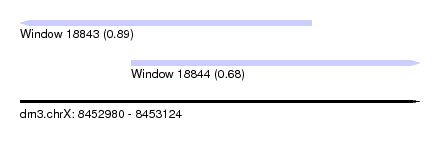
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,452,980 – 8,453,124 |
| Length | 144 |
| Max. P | 0.894803 |

| Location | 8,452,980 – 8,453,085 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.49393 |
| G+C content | 0.33194 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -9.02 |
| Energy contribution | -8.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8452980 105 - 22422827 -----------GACGAAAAGAAAACAUCGAUUUUUCACUAUACUAUCGAUGUUUCACGCCUUCAAAGU----GAAAUGUCCAAAAAAUAUGCCUCGUUUUCUUUAUAUGAUUACUAAAUG -----------.....(((((((((...((....))...........((((((((((.........))----))))))))...............)))))))))................ ( -19.30, z-score = -2.74, R) >droSec1.super_35 301572 88 - 471130 -----------GACGAAAAGAAAACAUCGAUUUCUCACUAUGCUAUCGAUGUUUCAGGUCUUCAAGGU---------------------UGCCUAGUUGUCUUUAUAUGAUUACUAAAUG -----------(((((...((((.(((((((..(.......)..)))))))))))((((..(....).---------------------.))))..)))))................... ( -18.50, z-score = -1.63, R) >droYak2.chrX 8911616 120 + 21770863 GACGAAAAGUGAGGAAAACAAAAACAUCGAAUCCUAGCUGUGCUAUCGAUGUUUCGCGUCUUUAAAGUUAUGUAUAUGUCCAACAAAUAUACCUCGUUUUCUUUAAAAGAUCAUUAUUUA ...((....((((((((((..(((((((((....((((...))))))))))))).................((((((.........))))))...)))))))))).....))........ ( -24.50, z-score = -2.17, R) >droEre2.scaffold_4690 12088969 116 + 18748788 GACGAAAAGUGAGCAAACCGAAAACAUCGAUUUCUAGCUAUGCAAUCGAUGUUUUGCGCCUUCAAAGU----UAUAUGUCCAACAUAUAUGCCUUGUUUUCUUUAUAAGAUCAUUAUUUA ...(((.((((((((...((((((((((((((.(.......).)))))))))))).))..........----((((((.....)))))))))(((((.......))))).))))).))). ( -25.20, z-score = -2.53, R) >consensus ___________AACAAAAAGAAAACAUCGAUUUCUAACUAUGCUAUCGAUGUUUCACGCCUUCAAAGU____UAUAUGUCCAACAAAUAUGCCUCGUUUUCUUUAUAAGAUCACUAAAUA .....................((((((((((..(.......)..)))))))))).................................................................. ( -9.02 = -8.90 + -0.12)

| Location | 8,453,020 – 8,453,124 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Shannon entropy | 0.42161 |
| G+C content | 0.38899 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8453020 104 + 22422827 ----GACAUUUCACUUUGAAGGCGUGAAACAUCGAUAGUAUAGUGAAAAAUCGAUGUUUU-----------CUUUUCGUCUUUUGUUUUGCUCUCGCUCUUCU-CCAUGAAGUGAUAACU ----......((((((((((((((.(((((((((((.............)))))))))))-----------.....)))))))......((....))......-....)))))))..... ( -24.52, z-score = -1.74, R) >droSec1.super_35 301603 94 + 471130 ------------ACCUUGAAGACCUGAAACAUCGAUAGCAUAGUGAGAAAUCGAUGUUUU-----------CUUUUCGUCUUUUGUUGUGCUCUCGGUCUUCU-UCCUAAAGUGAAAA-- ------------.....(((((((.((....))((.(((((((..(((...(((......-----------....)))..)))..))))))).))))))))).-..............-- ( -25.00, z-score = -2.78, R) >droYak2.chrX 8911656 120 - 21770863 GACAUAUACAUAACUUUAAAGACGCGAAACAUCGAUAGCACAGCUAGGAUUCGAUGUUUUUGUUUUCCUCACUUUUCGUCUUUUGUUUUGCUCUCGCUCUUCUGCCCUUUCGUGAAAACU ..(((...((.(((...(((((((.((((((((((((((...))))....))))))))))((.......)).....))))))).))).)).....((......))......)))...... ( -21.50, z-score = -0.64, R) >droEre2.scaffold_4690 12089009 113 - 18748788 ----GACAUAUAACUUUGAAGGCGCAAAACAUCGAUUGCAUAGCUAGAAAUCGAUGUUUUCGGUUUGCUCACUUUUCGUCUUUUGUUUUGCUCUCGCUCUUC---CCCUGCGUGAAAACU ----.............((((((((((((((.(((......(((...(((((((.....))))))))))......))).....))))))))....)).))))---......((....)). ( -23.90, z-score = -0.68, R) >consensus ____GACAUAUAACUUUGAAGACGCGAAACAUCGAUAGCAUAGCGAGAAAUCGAUGUUUU___________CUUUUCGUCUUUUGUUUUGCUCUCGCUCUUCU_CCCUUAAGUGAAAACU ............((...(((((((.(((((((((((..(.......)..)))))))))))................))))))).)).......((((..............))))..... (-14.27 = -14.02 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:06 2011