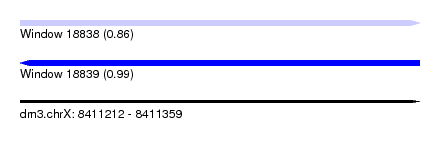
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,411,212 – 8,411,359 |
| Length | 147 |
| Max. P | 0.992016 |

| Location | 8,411,212 – 8,411,359 |
|---|---|
| Length | 147 |
| Sequences | 5 |
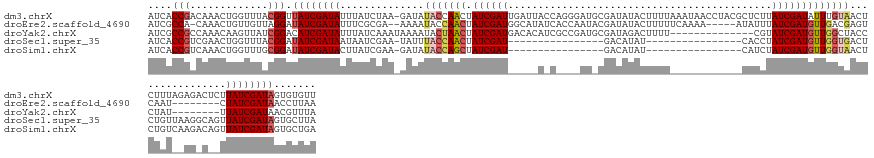
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 62.65 |
| Shannon entropy | 0.63831 |
| G+C content | 0.36741 |
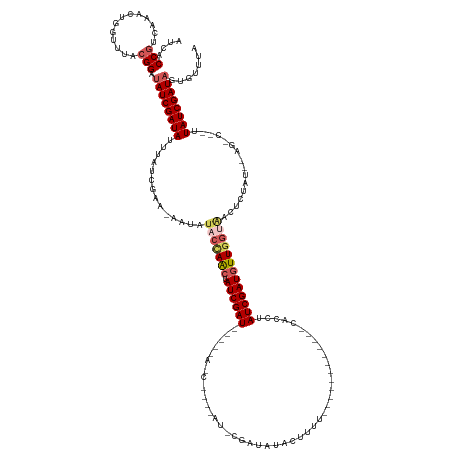
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
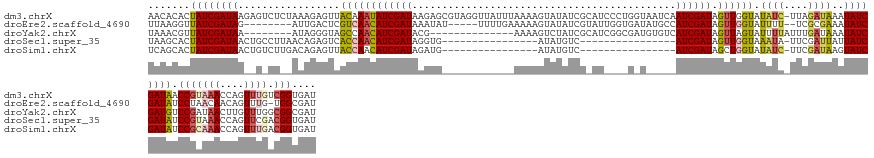
>dm3.chrX 8411212 147 + 22422827 AACACACUAUCGAUAAGAGUCUCUAAAGAGUUACAAAUAUCGAUAAGAGCGUAGGUUAUUUAAAAGUAUAUCGCAUCCCUGGUAAUCAAUCGAUAGUUGGUAUAUC-UUAGAUAAAUAUCGAUAACCGUAAACCAGUUUGUCGGUGAU .......((((((((...((((...((((..(((...(((((((....(((.....((((....))))...))).....((.....)))))))))....)))..))-))))))...))))))))((((((((....)))).))))... ( -29.80, z-score = -0.82, R) >droEre2.scaffold_4690 12047111 132 - 18748788 UUAAGGUUAUCGAUAG--------AUUGACUCGUCAACAUCGAUAAAUAU-----UUUUGAAAAAGUAUAUCGUAUUGGUGAUAUGCCAUCGAUAGUUGGUAUUUU--UCGCGAAAUAUCGAUAUCCUAACAACAGUUUG-UGGCGAU ...(((.((((((((.--------.((((....))))((.(((((..(((-----(((....)))))))))))...))((((.((((((.(....).))))))...--))))....)))))))).))).((((....)))-)...... ( -30.40, z-score = -0.99, R) >droYak2.chrX 8868408 126 - 21770863 UAAACGUUAUCGAUAA--------AUAGGGUAGCCAACAUCGAUACG--------------AAAAGUCUAUCGCAUCGGCGAUGUGUCAUCGAUAGUUAGUAUUUUAUUUGAUAAAUAUCGAUGUCCGAUAACUUGUUUGGCGGCGAU ((((((((((((....--------...((....)).(((((((((..--------------.......((((((....))))))(((((..(((((........))))))))))..))))))))).)))))))..)))))........ ( -30.90, z-score = -0.53, R) >droSec1.super_35 260305 115 + 471130 UAAGCACUAUCGAUAACUGCCUUAACAGAGUCACCAACAUCGAUAGGUG----------------AUAUGUC----------------AUCGAUAGUUGGUAAAUA-UUCGAUUAUUAUCGAUAUCCGUAAACCAGUUCGACGGUGAU ....(((..((((..((((..(((...((...((((((((((((.((..----------------.....))----------------)))))).)))))).....-.(((((....)))))..))..)))..))))))))..))).. ( -30.30, z-score = -2.21, R) >droSim1.chrX 6704607 115 + 17042790 UCAGCACUAUCGAUAACUGUCUUGACAGAGUUACCAACAUCGAUAGAUG----------------AUAUGUC----------------AUCGAUAGCUGGUAUAUC-UUCGAUAAGUAUCGAUAUCCGCAAACCAGUUUGACGGUGAU (((....((((((((.((...((((.(((..(((((.(((((((.(((.----------------....)))----------------)))))).).)))))..))-)))))..)))))))))).(((((((....)))).)))))). ( -33.60, z-score = -2.46, R) >consensus UAAACACUAUCGAUAA__G_CU__ACAGAGUCACCAACAUCGAUAAGUG____________AAAAGUAUAUCG_AU____G_U_____AUCGAUAGUUGGUAUAUC_UUCGAUAAAUAUCGAUAUCCGAAAACCAGUUUGACGGUGAU .......((((((((.................((((((((((((............................................)))))).)))))).((((....))))..)))))))).(((.............))).... (-13.26 = -13.22 + -0.04)
| Location | 8,411,212 – 8,411,359 |
|---|---|
| Length | 147 |
| Sequences | 5 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 62.65 |
| Shannon entropy | 0.63831 |
| G+C content | 0.36741 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -13.37 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.992016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
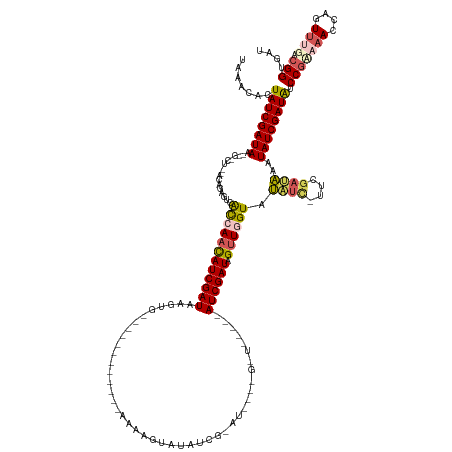
>dm3.chrX 8411212 147 - 22422827 AUCACCGACAAACUGGUUUACGGUUAUCGAUAUUUAUCUAA-GAUAUACCAACUAUCGAUUGAUUACCAGGGAUGCGAUAUACUUUUAAAUAACCUACGCUCUUAUCGAUAUUUGUAACUCUUUAGAGACUCUUAUCGAUAGUGUGUU .....((.((..(((((....(((((((((((..((((...-)))).......)))))).))))))))))...))))...................((((.((.(((((((...((..(((....)))))...))))))))).)))). ( -28.00, z-score = -0.54, R) >droEre2.scaffold_4690 12047111 132 + 18748788 AUCGCCA-CAAACUGUUGUUAGGAUAUCGAUAUUUCGCGA--AAAAUACCAACUAUCGAUGGCAUAUCACCAAUACGAUAUACUUUUUCAAAA-----AUAUUUAUCGAUGUUGACGAGUCAAU--------CUAUCGAUAACCUUAA ...((((-......((((.((......(((....)))...--....)).))))......))))(((((........)))))............-----....(((((((((((((....)))))--------..))))))))...... ( -23.92, z-score = -1.16, R) >droYak2.chrX 8868408 126 + 21770863 AUCGCCGCCAAACAAGUUAUCGGACAUCGAUAUUUAUCAAAUAAAAUACUAACUAUCGAUGACACAUCGCCGAUGCGAUAGACUUUU--------------CGUAUCGAUGUUGGCUACCCUAU--------UUAUCGAUAACGUUUA ...............((((((((.((((((((.(((.............))).)))))))).......((((((((((((((....)--------------).))))).)))))))........--------...))))))))..... ( -31.32, z-score = -2.75, R) >droSec1.super_35 260305 115 - 471130 AUCACCGUCGAACUGGUUUACGGAUAUCGAUAAUAAUCGAA-UAUUUACCAACUAUCGAU----------------GACAUAU----------------CACCUAUCGAUGUUGGUGACUCUGUUAAGGCAGUUAUCGAUAGUGCUUA ..(((..((((((((.((((((((..(((((....))))).-...((((((((.((((((----------------(......----------------....))))))))))))))).)))).)))).))))..))))..))).... ( -38.50, z-score = -4.36, R) >droSim1.chrX 6704607 115 - 17042790 AUCACCGUCAAACUGGUUUGCGGAUAUCGAUACUUAUCGAA-GAUAUACCAGCUAUCGAU----------------GACAUAU----------------CAUCUAUCGAUGUUGGUAACUCUGUCAAGACAGUUAUCGAUAGUGCUGA ..(((..((.(((((.((((((((..(((((....))))).-....(((((((.((((((----------------(......----------------....))))))))))))))..)))).)))).)))))...))..))).... ( -34.30, z-score = -2.36, R) >consensus AUCACCGUCAAACUGGUUUACGGAUAUCGAUAUUUAUCGAA_AAUAUACCAACUAUCGAU_____A_C____AU_CGAUAUACUUUU____________CACCUAUCGAUGUUGGUAACUCUAU__AG_C__UUAUCGAUAGUGUUUA ....(((.............))).((((((((..............(((((((.((((((............................................)))))))))))))................))))))))....... (-13.37 = -14.53 + 1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:26:01 2011