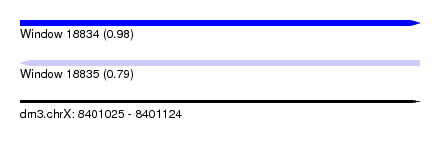
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,401,025 – 8,401,124 |
| Length | 99 |
| Max. P | 0.984305 |

| Location | 8,401,025 – 8,401,124 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Shannon entropy | 0.38333 |
| G+C content | 0.50028 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
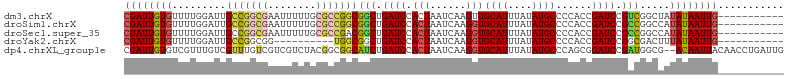
>dm3.chrX 8401025 99 + 22422827 CGAUUGUGUUUUGGAUUGCCGGCGAAUUUUUGCGCCGGCGGCUGAUCCACUAAUCAAGUUGCAUUUAUAUGCCCCACCGAUCCGUCGGCUAUAUAAUUG----------- .(((((((..(..(.(((((((((........))))))))))..)..))).)))).....((((....))))....((((....))))...........----------- ( -33.80, z-score = -2.63, R) >droSim1.chrX 6694536 99 + 17042790 CGAUUGUGUUUUGGAUUGCCGGCGAAUUUUUGCGCCGGCGGCUGAUCCACUAAUCAAGGUGCAUUUAUAUGCCCCACCGAUCCGCCGGCCAUAUAAUUG----------- .(((((((..(..(.(((((((((........))))))))))..)..))).))))..((.((((....)))).)).(((......)))...........----------- ( -36.40, z-score = -2.58, R) >droSec1.super_35 250364 99 + 471130 CGAUUGUGUUUUGGAUUGCCGGCGAAUUUUUGCGCCGACGGCUGAUCCACUAAUCAAGGUGCAUUUAUAUGCCCCACCGAUCCGCCGGCCAUAUAAUUG----------- ((((((((...(((.....(((((........))))).((((.((((.(((......)))((((....))))......)))).)))).)))))))))))----------- ( -30.00, z-score = -1.37, R) >droYak2.chrX 8857617 89 - 21770863 CGAUUGUGUUUUGGAUUGCCGGCGG----------UGGCGGCUGAUCCACUAAUCAAGGUGCAUUUAUAUGCCCCACCGAUCCGGCGACUUUAUAAUUG----------- ((((((((....((.((((((((((----------(((.(((.(((.((((......)))).))).....)))))))))..))))))))).))))))))----------- ( -36.60, z-score = -3.89, R) >dp4.chrXL_group1e 3463629 108 + 12523060 CGAUUGUGUCGUUUGUCGUUUGUCGUCGUCUACGGCGGCAUCUGAUCCACUAAUCAAGGUGCAUUUAUAUGCCCAGCGGAUCCGAUGGCG--ACAAUUACAACCUGAUUG .(.(((((....((((((((.(((((((....)))))))(((.(((((.((......((.((((....)))))))).))))).)))))))--)))).))))).)...... ( -37.30, z-score = -2.60, R) >consensus CGAUUGUGUUUUGGAUUGCCGGCGAAUUUUUGCGCCGGCGGCUGAUCCACUAAUCAAGGUGCAUUUAUAUGCCCCACCGAUCCGCCGGCCAUAUAAUUG___________ ((((((((.........(((((((........)))))))(((.((((.(((......)))((((....))))......)))).))).....))))))))........... (-20.82 = -21.70 + 0.88)
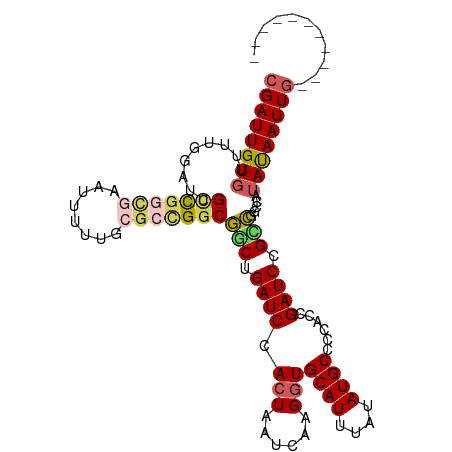
| Location | 8,401,025 – 8,401,124 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Shannon entropy | 0.38333 |
| G+C content | 0.50028 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -17.70 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8401025 99 - 22422827 -----------CAAUUAUAUAGCCGACGGAUCGGUGGGGCAUAUAAAUGCAACUUGAUUAGUGGAUCAGCCGCCGGCGCAAAAAUUCGCCGGCAAUCCAAAACACAAUCG -----------................((((.((((..((((....))))..).(((((....))))))))(((((((........))))))).))))............ ( -29.60, z-score = -1.81, R) >droSim1.chrX 6694536 99 - 17042790 -----------CAAUUAUAUGGCCGGCGGAUCGGUGGGGCAUAUAAAUGCACCUUGAUUAGUGGAUCAGCCGCCGGCGCAAAAAUUCGCCGGCAAUCCAAAACACAAUCG -----------..........((((((((((((((((.((((....))))...((((((....))))))))))))........))))))))))................. ( -34.20, z-score = -1.97, R) >droSec1.super_35 250364 99 - 471130 -----------CAAUUAUAUGGCCGGCGGAUCGGUGGGGCAUAUAAAUGCACCUUGAUUAGUGGAUCAGCCGUCGGCGCAAAAAUUCGCCGGCAAUCCAAAACACAAUCG -----------..........(((((((((((((((((((((....)))).)))(((((....)))))))))...........))))))))))................. ( -31.00, z-score = -1.36, R) >droYak2.chrX 8857617 89 + 21770863 -----------CAAUUAUAAAGUCGCCGGAUCGGUGGGGCAUAUAAAUGCACCUUGAUUAGUGGAUCAGCCGCCA----------CCGCCGGCAAUCCAAAACACAAUCG -----------.............(((((..(((((((((((....)))).)).......((((.....))))))----------))))))))................. ( -28.00, z-score = -2.13, R) >dp4.chrXL_group1e 3463629 108 - 12523060 CAAUCAGGUUGUAAUUGU--CGCCAUCGGAUCCGCUGGGCAUAUAAAUGCACCUUGAUUAGUGGAUCAGAUGCCGCCGUAGACGACGACAAACGACAAACGACACAAUCG .......(((((..((((--((.((((.((((((((((((((....)))).))......)))))))).)))).((.((....)).)).....)))))))))))....... ( -33.90, z-score = -2.48, R) >consensus ___________CAAUUAUAUAGCCGGCGGAUCGGUGGGGCAUAUAAAUGCACCUUGAUUAGUGGAUCAGCCGCCGGCGCAAAAAUUCGCCGGCAAUCCAAAACACAAUCG .....................((((.(((((((((((.((((....))))...((((((....))))))))))))........))))).))))................. (-17.70 = -19.30 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:58 2011