| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,392,036 – 8,392,140 |
| Length | 104 |
| Max. P | 0.766694 |

| Location | 8,392,036 – 8,392,140 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Shannon entropy | 0.22540 |
| G+C content | 0.49474 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -22.92 |
| Energy contribution | -24.96 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8392036 104 + 22422827 GAUUCUGCCAAUUUCGAUCGAUCGAUCGAUAACCAUUAUCUCGCCCAGUGGCCUCUUAAACGGCUUUCAAUGCUCAGCCGAGUGGUACUGGUUAGUGGGAUCAG-------- (((((..(.....((((((....))))))((((((.((((.((((....)))........(((((..........))))).).)))).)))))))..)))))..-------- ( -36.10, z-score = -3.16, R) >droSim1.chrX 6684717 98 + 17042790 GAUUCUGCCAAUUUCGAUCGAUCGAUCGAUAACCAUUAUCUCGCCCAGUGGCCUCUUAAACGGCUUUCAAUGCUCAGCCGA------CUGGUUAGCGGGAUCAG-------- ((((((((.....((((((....))))))((((((.......(((....)))........(((((..........))))).------.))))))))))))))..-------- ( -34.10, z-score = -3.42, R) >droSec1.super_35 241448 103 + 471130 GAUUCUGCCAAUUUCGAUCGAUCGAUCGAUAACCAUUAUCUCGCCCAGUGGCCUCUUAAACGGCUUUCAAUGCUCAGCCGACUGGU-UAGGUUAGCGGGAUCAG-------- ((((((((.((((((((((....))))))((((((.......(((....)))........(((((..........)))))..))))-)))))).))))))))..-------- ( -34.70, z-score = -2.87, R) >droYak2.chrX 8848055 106 - 21770863 GAUUCUGGCAGCUUCGAUCGAUCGAUCGAUAACCAUUAUCUCGCCCAGUGGCCUCUUGAACGACAUUCAAUGCUCAGCCGA------CUGGUUAGCGGGAUCAGAUCGGAAU .((((((((....((((((....))))))........(((((((((((((((...(((((.....)))))......))).)------))))...)))))))..).))))))) ( -33.10, z-score = -0.92, R) >droEre2.scaffold_4690 12027942 94 - 18748788 GAUUCUGCCAGUUUCGAUGGAUUGAU----AACCAUUAUCUCGCCCAGUAGCCUCUUAAACGACUUUCAAUGCUCAGCCGA------CUGGUCAGCGGGAUCAA-------- ((((((((.......(((((......----..))))).....(.(((((.((........................))..)------)))).).))))))))..-------- ( -20.96, z-score = 0.00, R) >consensus GAUUCUGCCAAUUUCGAUCGAUCGAUCGAUAACCAUUAUCUCGCCCAGUGGCCUCUUAAACGGCUUUCAAUGCUCAGCCGA______CUGGUUAGCGGGAUCAG________ ((((((((.....((((((....))))))(((((........(((....)))........(((((..........))))).........))))))))))))).......... (-22.92 = -24.96 + 2.04)


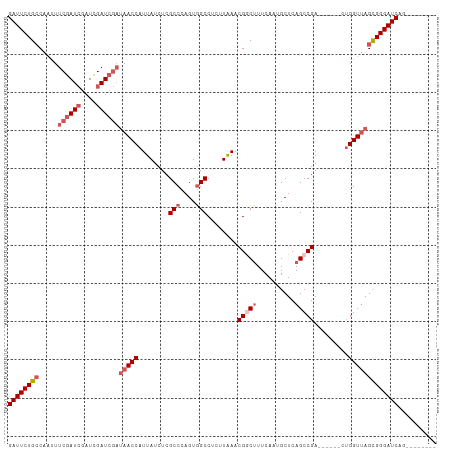
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:55 2011