| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,376,948 – 8,377,044 |
| Length | 96 |
| Max. P | 0.612913 |

| Location | 8,376,948 – 8,377,044 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.67 |
| Shannon entropy | 0.53957 |
| G+C content | 0.65097 |
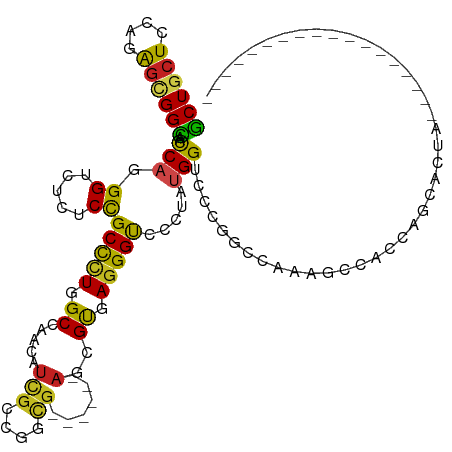
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -16.46 |
| Energy contribution | -15.13 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8376948 96 + 22422827 GCUGCUCCAGAGCGGCAACCAGGGUCUGUCCGCUCUUGCCAACAUCGCCGGCG------AGCGUGAGGGUCCCUAUGGUCCCGCUCCAAGCCAUCAGCACUA------------------ ((((((...((((((..(((((((.((.(((((((..(((.........))))------)))).)).)).)))..)))).))))))..)))....)))....------------------ ( -41.50, z-score = -2.44, R) >droSim1.chrX_random 2512557 96 + 5698898 GCUGCUCCAGAGCGGCAACCAGGGUCUGUCCGCUCUCGCCAACAUCGCUGGCG------AGCGUGAGGGUCCCUAUGGUCCCGCUCCCAGCCAUCAGCACUA------------------ ((((((...((((((..(((((((.((.(((((((..((((.......)))))------)))).)).)).)))..)))).))))))..)))....)))....------------------ ( -43.10, z-score = -2.69, R) >droSec1.super_35 225818 96 + 471130 GCUGCUCCAGAGCGGCAACCAGGGUCUGUCCGCUCUCGCCAACAUUGCCGGCG------AGCGUGAGGGUCCCUAUGGUCCCGCUCCCAGCCAUCAGCACUA------------------ ((((((...((((((..(((((((.((.(((((((..(((.........))))------)))).)).)).)))..)))).))))))..)))....)))....------------------ ( -41.50, z-score = -2.29, R) >droYak2.chrX 8833523 96 - 21770863 GCUGCUCCAGAGCGGCAACCAGGGUCUGUCCGCUCUCGCCAACAUCGCCGGCG------AGCGUGAGGGUCCCUAUGGUCCAGCCCCAAGCCAUCAGCACUA------------------ ((((((....)))(((....((((.((.(((((((..(((.........))))------)))).)).)).)))).(((.......))).)))...)))....------------------ ( -36.60, z-score = -1.02, R) >droEre2.scaffold_4690 12013340 96 - 18748788 GCUGCUCCAGAGCGGUAACCAGGGUCUGUCCGCCCUCGCCAGCAUCGCCGGCG------AGCGUGAGGGUCCCUAUGGUCCUGGCCCAAGCCAGCAGCACUA------------------ (((((........((..(((((((.((.(((((..(((((.((...)).))))------)))).)).)).)))..))))))((((....)))))))))....------------------ ( -42.90, z-score = -1.50, R) >droAna3.scaffold_13248 2303656 96 - 4840945 GCUCCUCCAGAACGGCAACCAGGGCCUCUCCGCCCUGGCCAGCAUCGCCGGCG------AUCGUGAGGGUCCCUAUGGUGCCGGACCUAGCCACCAGAGCUA------------------ (((((....)...(((..(((((((......)))))))((.((((((..((.(------(((.....))))))..)))))).)).....)))....))))..------------------ ( -39.20, z-score = -0.77, R) >dp4.chrXL_group1e 3438067 114 + 12523060 GCUGCUCCAGGGUGGCAAUCAGGGUCUCUCCGCCCUGGCCAGCAUCGCUGGCG------AUCGCGAGGGUCACUACGGUGGCGGUCACGAUCACGGUCACGGUCACAGCCAUCAGCACUA ((((.......(((((...((((((......))))))((((((...))))))(------((((.((..(((((....)))))..)).)))))...)))))(((....)))..)))).... ( -53.00, z-score = -2.92, R) >droPer1.super_17 375050 114 + 1930428 GCUGCUCCAGGGUGGCAAUCAGGGUCUCUCCGCCCUGGCCAGCAUCGCUGGCG------AUCGCGAGGGUCACUACGGUGGCGGUCACGAUCACGGUCACGGUCACAGCCAUCAGCACUA ((((.......(((((...((((((......))))))((((((...))))))(------((((.((..(((((....)))))..)).)))))...)))))(((....)))..)))).... ( -53.00, z-score = -2.92, R) >droWil1.scaffold_180702 1102603 96 + 4511350 ACUCCUCCAGGGUGGUAACCACGGCCUCUCUGCCUUAGCCAACAUUGCCGGUGGUGAGCACCGUGAGGGUCACUAUGGUGGUGCCCAGCAAUACGG------------------------ ...((..(((((.(((.......))).))))).....((...(((..(((.((((((.(.((....))))))))))))..)))....)).....))------------------------ ( -32.50, z-score = 0.81, R) >droVir3.scaffold_12970 890894 84 - 11907090 GCUGCUCCAGGGUGGCAACCAUGGUCUCUCCGCUCUGGCCAACAUUGCCGGCG------AGCGUGAAGGUAACUAUGG---UAGCCAGAGCUA--------------------------- ...((((.....((((.(((((((((..(((((((((((.......))))..)------)))).))..)..)))))))---).))))))))..--------------------------- ( -35.70, z-score = -1.78, R) >droMoj3.scaffold_6473 10331268 84 + 16943266 GCUGCUCCAGGGCGGCAACCAUGGUCUCUCCGCCCUGGCCAACAUUGCUGGCG------ACCGCGAGGGCAGCUAUGG---CGGCCAGAGCUA--------------------------- ...((((.(((((((..((....))....)))))))((((..(((.((((...------.((....)).)))).))).---.)))).))))..--------------------------- ( -41.10, z-score = -1.43, R) >droGri2.scaffold_15203 8986627 81 - 11997470 GCUGCUCCAGGGUGGCAACCAUGGACUCUCUGCUCUGGCCAACAUCGCCGGA---------CGUGAGGGCAGCUAUGG---CGGCCAAGGCGG--------------------------- .(((((......((((..((((((.(((((.((((((((.......))))))---------.)))))))...))))))---..)))).)))))--------------------------- ( -35.80, z-score = -1.08, R) >consensus GCUGCUCCAGAGCGGCAACCAGGGUCUCUCCGCCCUGGCCAACAUCGCCGGCG______AGCGUGAGGGUCCCUAUGGUCCCGGCCAAAGCCACCAGCACUA__________________ ((((((....)))))).....(((.....(((.....(((....((((..............)))).))).....)))......)))................................. (-16.46 = -15.13 + -1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:51 2011