
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,315,810 – 8,315,929 |
| Length | 119 |
| Max. P | 0.898035 |

| Location | 8,315,810 – 8,315,929 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Shannon entropy | 0.44059 |
| G+C content | 0.37881 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.84 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8315810 119 + 22422827 UCGUAAUCGUUUUUAGGGCAAACU-GUUUAUCCAUAAAAAAUGUGGGCUUUGUAAAUAGCAUCAAUUCGCUUUUUUGUGGGUCAUCCAAAAAGAAACCGAAACACCCACACACACCCACA .........((((((((..((...-..))..)).)))))).((((((...((((((.(((........))).))).((((((..((............))...)))))))))..)))))) ( -22.40, z-score = -0.20, R) >droEre2.scaffold_4690 11956190 109 - 18748788 UAGCGAUCGUUUUAGGUGUAAACUGAUUAUCCAGAGAAAAGUGUGGGCUUUUUACAUAGCAUCAAUUCGCUUUUU-GUAGGUCAUCCAAACAGAAACCAAAACACCCACA---------- ..............(((((...(((......)))......((.((((...((((((.(((........)))...)-)))))...)))).))..........)))))....---------- ( -20.60, z-score = -0.42, R) >droYak2.chrX 8770905 119 - 21770863 UAGUAAUUGUUUUUAGAGCAAACAUAUUAUCCAGGAAAAAGUGUGGGCCUUUUGAAUAGCAUCAAUUCGCUUUUU-GUGGGUCAUCCAAAAAGAAACCAAAACACCCACACACACCCACA ..((((((((((.......))))).)))))...((.....(((((((..(((((....((........))(((((-.(((.....))).)))))...)))))..)))))))....))... ( -23.60, z-score = -0.85, R) >droSec1.super_35 165291 110 + 471130 UCUUAAUCGUUUUAAGUUUAAACU-A---------AAAAAACGUGGGCCUUUUAAAUAGCAUCUAUUCGCUUUUUUGUGGGUCAUCCAAAAAGAAACCAAAACACCCACACACACCCACA ........(((((.(((....)))-.---------..)))))(((((......(((.(((........))).)))(((((((.....................)))))))....))))). ( -20.70, z-score = -2.32, R) >droSim1.chrX_random 2494456 95 + 5698898 ---------------UCGUAAACU-A---------AAAAAACGUGGGCUUUUCAAAUAGCAUCAAUUCGCUUUUUUGUGGGUCAUCCAAAAAGAAACCAAAACACCCACACACACCCACA ---------------.........-.---------.......(((((......(((.(((........))).)))(((((((.....................)))))))....))))). ( -19.00, z-score = -2.42, R) >consensus UAGUAAUCGUUUUAAGUGUAAACU_AUU___C___AAAAAAUGUGGGCUUUUUAAAUAGCAUCAAUUCGCUUUUUUGUGGGUCAUCCAAAAAGAAACCAAAACACCCACACACACCCACA ..........................................(((((..........(((........))).....((((((.....................)))))).....))))). (-12.64 = -13.84 + 1.20)



| Location | 8,315,810 – 8,315,929 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Shannon entropy | 0.44059 |
| G+C content | 0.37881 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8315810 119 - 22422827 UGUGGGUGUGUGUGGGUGUUUCGGUUUCUUUUUGGAUGACCCACAAAAAAGCGAAUUGAUGCUAUUUACAAAGCCCACAUUUUUUAUGGAUAAAC-AGUUUGCCCUAAAAACGAUUACGA (((((((...(((((((((.(((((((((((((((.....)))...))))).))))))).)).)))))))..))))))).((((((.((.(((..-...))))).))))))......... ( -31.00, z-score = -1.28, R) >droEre2.scaffold_4690 11956190 109 + 18748788 ----------UGUGGGUGUUUUGGUUUCUGUUUGGAUGACCUAC-AAAAAGCGAAUUGAUGCUAUGUAAAAAGCCCACACUUUUCUCUGGAUAAUCAGUUUACACCUAAAACGAUCGCUA ----------(((((((..((..(.......)..))..))))))-)...(((((..((((.(((.(..(((((......)))))..))))...))))((((.......))))..))))). ( -24.00, z-score = -0.86, R) >droYak2.chrX 8770905 119 + 21770863 UGUGGGUGUGUGUGGGUGUUUUGGUUUCUUUUUGGAUGACCCAC-AAAAAGCGAAUUGAUGCUAUUCAAAAGGCCCACACUUUUUCCUGGAUAAUAUGUUUGCUCUAAAAACAAUUACUA ...(((...((((((((.((((((....(((.(((.....))).-))).((((......))))..)))))).))))))))....)))(((.((((.(((((.......)))))))))))) ( -30.80, z-score = -1.41, R) >droSec1.super_35 165291 110 - 471130 UGUGGGUGUGUGUGGGUGUUUUGGUUUCUUUUUGGAUGACCCACAAAAAAGCGAAUAGAUGCUAUUUAAAAGGCCCACGUUUUUU---------U-AGUUUAAACUUAAAACGAUUAAGA .((((((.(.(((((((..((..(.......)..))..)))))))(((.((((......)))).)))...).))))))(((((..---------.-(((....))).)))))........ ( -29.60, z-score = -2.26, R) >droSim1.chrX_random 2494456 95 - 5698898 UGUGGGUGUGUGUGGGUGUUUUGGUUUCUUUUUGGAUGACCCACAAAAAAGCGAAUUGAUGCUAUUUGAAAAGCCCACGUUUUUU---------U-AGUUUACGA--------------- .((((((...(((((((..((..(.......)..))..)))))))(((.((((......)))).))).....)))))).......---------.-.........--------------- ( -26.00, z-score = -1.87, R) >consensus UGUGGGUGUGUGUGGGUGUUUUGGUUUCUUUUUGGAUGACCCACAAAAAAGCGAAUUGAUGCUAUUUAAAAAGCCCACAUUUUUU___G___AAU_AGUUUACACUAAAAACGAUUACGA ..........(((((((.(((((((((((....))).))))........((((......))))....)))).)))))))......................................... (-18.62 = -18.66 + 0.04)

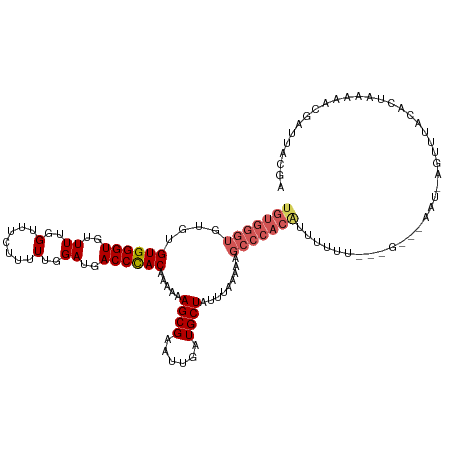
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:49 2011