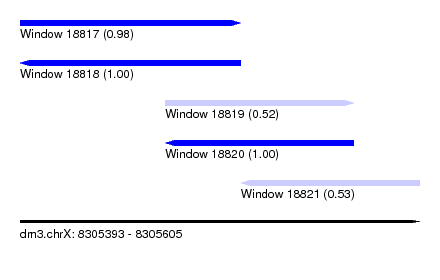
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,305,393 – 8,305,605 |
| Length | 212 |
| Max. P | 0.997994 |

| Location | 8,305,393 – 8,305,510 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.42242 |
| G+C content | 0.43496 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8305393 117 + 22422827 GUAUCGUUGGUUUACAAUGGGCACAUUGGAACACUCUUCUAUGGAAAGUGCACCACUUGC---CCACUGUGUUUUUUCUCUCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGC ....((..((..((((.((((((...(((..((((.(((....)))))))..)))..)))---))).)))).................(((((....)))))..........))...)). ( -30.70, z-score = -2.47, R) >droEre2.scaffold_4690 11945831 113 - 18748788 --AUCGUUGGUUUACAAUGAGC--AUUAGAACACACUCCUAUGGAUAGUGCUUAUUUUGC---UCACUGUAUUUUUCUCCGUUUUUUGGCUGCCCCAGCGGGAAAUCUUUAUCCUUUCGC --..((((((..((((.(((((--(....(((((..(((...)))..))).))....)))---))).)))).......(((.....))).....))))))(((........)))...... ( -26.60, z-score = -1.62, R) >droYak2.chrX 8761902 94 - 21770863 UUAUCGUUGGUUUACAAUGGGC--AUGGAAAUACUCUUCUAUGAAAAGUGCUUCUUUUGC---CCACUGUGUUUUUCUCCAUUUUUCGAAUCCUUUCGA--------------------- .......(((..((((.(((((--(.((((.((((.(((...))).)))).))))..)))---))).)))).......)))....(((((....)))))--------------------- ( -24.80, z-score = -3.75, R) >droSec1.super_35 155681 120 + 471130 GUAUCGUUGGUUUACAAUGGGCACAUUGGAGCACUUUUCUAUGGAAAGUGCACCACUUGCCGCCCAAUGUGUUUUUUCUCUCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGC ....((..((..((((.(((((.((.(((.((((((((.....)))))))).)))..))..))))).)))).................(((((....)))))..........))...)). ( -36.70, z-score = -3.41, R) >droSim1.chrX 6611356 120 + 17042790 GUAUCGUUGGUUUACAAUGGGCACAUUGGAACACUUUUCUAUGGAAAGUGCACCACUUGCCGCCCAAUGUGUUUUUUCUCUCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGC ....((..((..((((.(((((.((.(((..(((((((.....)))))))..)))..))..))))).)))).................(((((....)))))..........))...)). ( -32.50, z-score = -2.47, R) >consensus GUAUCGUUGGUUUACAAUGGGCACAUUGGAACACUCUUCUAUGGAAAGUGCACCACUUGC___CCACUGUGUUUUUUCUCUCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGC ............((((.((((((...(((..((((.(((...))).))))..)))..)))...))).)))).................(((((....))))).................. (-14.94 = -15.34 + 0.40)

| Location | 8,305,393 – 8,305,510 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.42242 |
| G+C content | 0.43496 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -18.26 |
| Energy contribution | -21.06 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8305393 117 - 22422827 GCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGAGAGAAAAAACACAGUGG---GCAAGUGGUGCACUUUCCAUAGAAGAGUGUUCCAAUGUGCCCAUUGUAAACCAACGAUAC ......((.......(((((.((....)).)))))...............(((((((---(((..(((.(((((((.......))))))).)))...))))))))))...))........ ( -37.90, z-score = -3.85, R) >droEre2.scaffold_4690 11945831 113 + 18748788 GCGAAAGGAUAAAGAUUUCCCGCUGGGGCAGCCAAAAAACGGAGAAAAAUACAGUGA---GCAAAAUAAGCACUAUCCAUAGGAGUGUGUUCUAAU--GCUCAUUGUAAACCAACGAU-- ......((.......(((((((.(((.....))).....))).))))..((((((((---(((.....(((((..(((...)))..)))))....)--))))))))))..))......-- ( -30.30, z-score = -2.72, R) >droYak2.chrX 8761902 94 + 21770863 ---------------------UCGAAAGGAUUCGAAAAAUGGAGAAAAACACAGUGG---GCAAAAGAAGCACUUUUCAUAGAAGAGUAUUUCCAU--GCCCAUUGUAAACCAACGAUAA ---------------------(((((....)))))....(((.(.....)(((((((---(((...((((.(((((((...))))))).))))..)--)))))))))...)))....... ( -27.50, z-score = -4.98, R) >droSec1.super_35 155681 120 - 471130 GCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGAGAGAAAAAACACAUUGGGCGGCAAGUGGUGCACUUUCCAUAGAAAAGUGCUCCAAUGUGCCCAUUGUAAACCAACGAUAC ......((.......(((((.((....)).)))))...............(((.((((((.((..(((.(((((((.......))))))).))).)))))))).)))...))........ ( -35.40, z-score = -2.23, R) >droSim1.chrX 6611356 120 - 17042790 GCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGAGAGAAAAAACACAUUGGGCGGCAAGUGGUGCACUUUCCAUAGAAAAGUGUUCCAAUGUGCCCAUUGUAAACCAACGAUAC ......((.......(((((.((....)).)))))...............(((.((((((.((..(((.(((((((.......))))))).))).)))))))).)))...))........ ( -33.10, z-score = -1.64, R) >consensus GCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGAGAGAAAAAACACAGUGG___GCAAGUGGUGCACUUUCCAUAGAAGAGUGUUCCAAUGUGCCCAUUGUAAACCAACGAUAC ......((.......(((((.((....)).)))))...............(((((((...(((..(((.(((((((.......))))))).)))...))))))))))...))........ (-18.26 = -21.06 + 2.80)

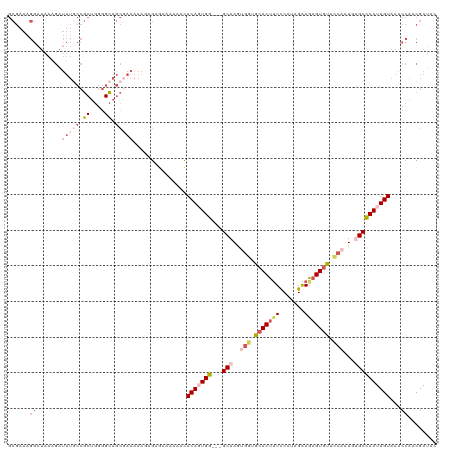
| Location | 8,305,470 – 8,305,570 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.76 |
| Shannon entropy | 0.21115 |
| G+C content | 0.52375 |
| Mean single sequence MFE | -36.29 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8305470 100 + 22422827 UCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGCAUCGAAUUCGCCAUCUAACAGUGUGCGUUAACAUG-------AAAGCGCUAACAGCGGCAGGGC-AGC------------ ........(((((((..((.((.............((((...))))..............((..(((((......-------..)))))..)).)).)).))))-)))------------ ( -27.60, z-score = -0.72, R) >droEre2.scaffold_4690 11945904 120 - 18748788 GUUUUUUGGCUGCCCCAGCGGGAAAUCUUUAUCCUUUCGCAUCAAAUUCGCCCUCUAACAGUCUGCGUUAACAUGCUGUUAGAAAGCGCUAACAGUAGCGGGGCGAGCGACAGGGGCAGC ........((((((((.(((..(...........)..)))......((((((((((((((((.((......)).)))))))))...(((((....)))))))))))).....)))))))) ( -53.10, z-score = -4.79, R) >droSec1.super_35 155761 100 + 471130 UCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGCAUCAAAUUCGCCCUCUAACAGUGCGCGUUAACAUG-------AAAGCGCUAACAGCGGCGGGGC-AGC------------ ........((((((((.((((...............)))).........(((((.....(((((.(((....)))-------...)))))...)).))))))))-)))------------ ( -32.86, z-score = -2.04, R) >droSim1.chrX 6611436 100 + 17042790 UCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGCAUCGAAUUCGCCCUCUAACAGUGUGCGUUAACAUG-------AAAGCGCUAACAGCGGCGGGGC-AGC------------ ........((((((((.((.((.............((((...))))..............((..(((((......-------..)))))..)).)).)))))))-)))------------ ( -31.60, z-score = -1.39, R) >consensus UCUUUUUCGCUGCCCCAGCGGCAAAUCUUUAUCCUUUCGCAUCAAAUUCGCCCUCUAACAGUGUGCGUUAACAUG_______AAAGCGCUAACAGCGGCGGGGC_AGC____________ ........((((((((.((.((................((.........)).........((..(((((...............)))))..)).)).))))))).)))............ (-22.57 = -22.89 + 0.31)

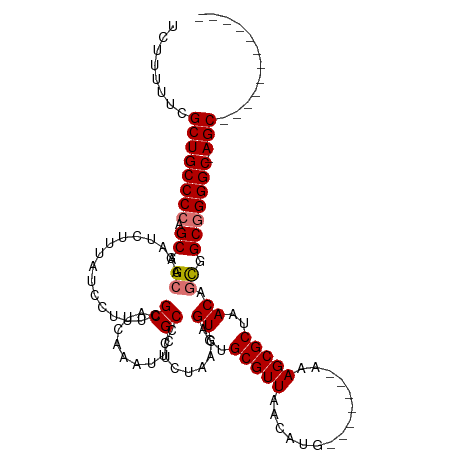
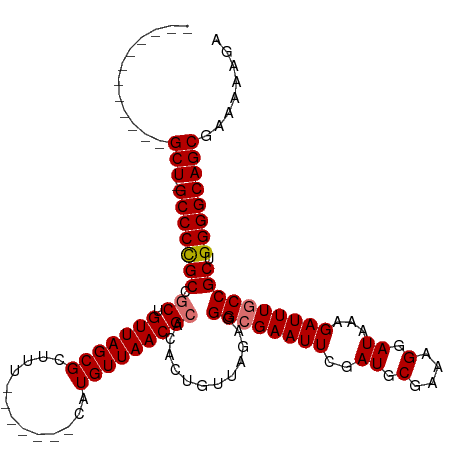
| Location | 8,305,470 – 8,305,570 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.76 |
| Shannon entropy | 0.21115 |
| G+C content | 0.52375 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -33.43 |
| Energy contribution | -33.74 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.997994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8305470 100 - 22422827 ------------GCU-GCCCUGCCGCUGUUAGCGCUUU-------CAUGUUAACGCACACUGUUAGAUGGCGAAUUCGAUGCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGA ------------(((-(((((((.((.(((((((....-------..)))))))))............((((((((..((.(....).))...)))))))))).))))))))........ ( -40.00, z-score = -3.27, R) >droEre2.scaffold_4690 11945904 120 + 18748788 GCUGCCCCUGUCGCUCGCCCCGCUACUGUUAGCGCUUUCUAACAGCAUGUUAACGCAGACUGUUAGAGGGCGAAUUUGAUGCGAAAGGAUAAAGAUUUCCCGCUGGGGCAGCCAAAAAAC ((((((((.((((.(((((((((((....)))))...((((((((..(((....)))..))))))))))))))...))))(((...(((........)))))).))))))))........ ( -52.60, z-score = -5.25, R) >droSec1.super_35 155761 100 - 471130 ------------GCU-GCCCCGCCGCUGUUAGCGCUUU-------CAUGUUAACGCGCACUGUUAGAGGGCGAAUUUGAUGCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGA ------------(((-((((((((((.(((((((....-------..))))))))))...........(((((((((.((.(....).))..))))))))))).))))))))........ ( -43.30, z-score = -3.50, R) >droSim1.chrX 6611436 100 - 17042790 ------------GCU-GCCCCGCCGCUGUUAGCGCUUU-------CAUGUUAACGCACACUGUUAGAGGGCGAAUUCGAUGCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGA ------------(((-(((((((.((.(((((((....-------..)))))))))............((((((((..((.(....).))...)))))))))).))))))))........ ( -42.20, z-score = -3.44, R) >consensus ____________GCU_GCCCCGCCGCUGUUAGCGCUUU_______CAUGUUAACGCACACUGUUAGAGGGCGAAUUCGAUGCGAAAGGAUAAAGAUUUGCCGCUGGGGCAGCGAAAAAGA ............(((.(((((((.((.(((((((.............)))))))))............((((((((..((.(....).))...)))))))))).))))))))........ (-33.43 = -33.74 + 0.31)

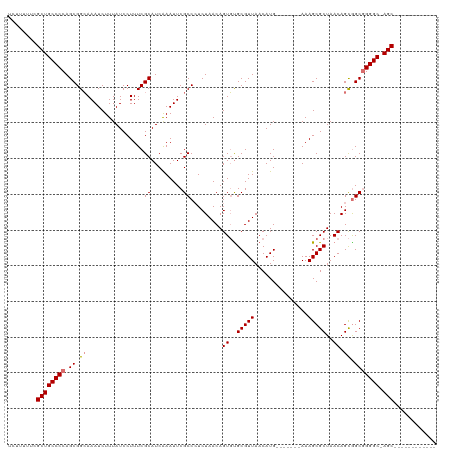
| Location | 8,305,510 – 8,305,605 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.60 |
| Shannon entropy | 0.34106 |
| G+C content | 0.55730 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.35 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8305510 95 - 22422827 AACUCUCC----CCCUCACCCUUUCGCCUUGUGGCUCUGGCU-------------GCCCUGCCGCUGUUAGCGCUUU-------CAUGUUAACGCACACUGUUAGAUGGCGAAUUCGAU ........----..........((((((.(.((((...(((.-------------.....)))((.(((((((....-------..))))))))).....)))).).))))))...... ( -25.20, z-score = -1.72, R) >droEre2.scaffold_4690 11945944 118 + 18748788 AGCUCUCUCUCCCCCUCCCCCUUUCGCCAUGU-UCUCUGGCUGCCCCUGUCGCUCGCCCCGCUACUGUUAGCGCUUUCUAACAGCAUGUUAACGCAGACUGUUAGAGGGCGAAUUUGAU .........................((((...-....)))).......((((.(((((((((((....)))))...((((((((..(((....)))..))))))))))))))...)))) ( -33.30, z-score = -2.16, R) >droYak2.chrX 8761996 115 + 21770863 AGCUCUC---CCCCCACCCCCUAUCAUCAUGC-UCUCUGGCUGCCCCUGCCGUUCGCCCCGCUACUGUUAGCGCUUUCUAACAGCAUGUUAACGCAGACUGUUAGAGGGCGCAUUUGAU .......---............((((..((((-.....(((.......)))....(((((((((....)))))...((((((((..(((....)))..)))))))))))))))).)))) ( -35.10, z-score = -2.94, R) >droSec1.super_35 155801 96 - 471130 AACUCUCC--CUCCCUCACCCUUUCGCUUUGU-UCUCUGGCU-------------GCCCCGCCGCUGUUAGCGCUUU-------CAUGUUAACGCGCACUGUUAGAGGGCGAAUUUGAU ........--.....((.((((((.((..(((-.....(((.-------------.....)))((.(((((((....-------..))))))))))))..)).)))))).))....... ( -25.60, z-score = -1.65, R) >droSim1.chrX 6611476 96 - 17042790 AACUCUCC--CCCCCUCCCCCUUUCGCCUUGU-UCUCUGGCU-------------GCCCCGCCGCUGUUAGCGCUUU-------CAUGUUAACGCACACUGUUAGAGGGCGAAUUCGAU ........--.............(((..((((-((((((((.-------------.....)))((.(((((((....-------..)))))))))........)))))))))...))). ( -23.70, z-score = -1.63, R) >consensus AACUCUCC__CCCCCUCCCCCUUUCGCCUUGU_UCUCUGGCU_____________GCCCCGCCGCUGUUAGCGCUUU_______CAUGUUAACGCACACUGUUAGAGGGCGAAUUUGAU ...............((.((((((.((..(((.....((((...................))))..(((((((.............))))))))))....)).)))))).))....... (-14.87 = -14.35 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:46 2011