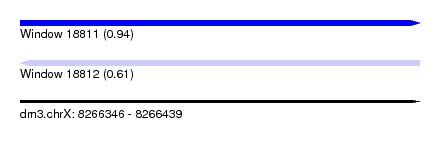
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,266,346 – 8,266,439 |
| Length | 93 |
| Max. P | 0.943587 |

| Location | 8,266,346 – 8,266,439 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.53529 |
| G+C content | 0.46451 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -11.57 |
| Energy contribution | -12.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
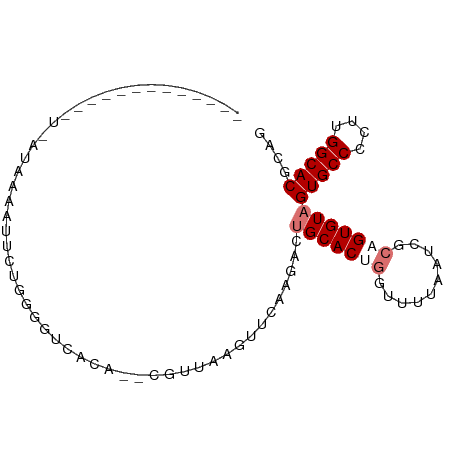
>dm3.chrX 8266346 93 + 22422827 GUUUAAUGCACUUUCAUAAAGUUCGGGGGUCACAUACGCUUAGUUUAAGACUGCACUGGUUUUAAGCGCUGUGUAGUGCCCCUUGGCACGCAG ......(((((((.....))))(((((((.(((.(((((...((((((((((.....))))))))))...)))))))).)))))))...))). ( -32.50, z-score = -2.74, R) >droEre2.scaffold_4690 10631797 79 + 18748788 -------------UGAUAACCUU-UUGGGUCAGAUGUGUGAAGUUCUAGAGCGCACUGGUUUUAAUCCCAGUGUAGUGCCCCUUGGCACGCAG -------------((((..((..-..))))))..................(((((((((........))))))).(((((....))))))).. ( -23.60, z-score = -1.39, R) >droYak2.chrX 8723158 76 - 21770863 ---------------AUGAUACCCUGAAUCUAAA--CAUUAGUUUCAGGACUGCACUCGCUUUAAUCGCAGUGUAGUGCCCCUUGGCACGCAG ---------------.......((((((.(((..--...))).))))))((((((((.((.......))))))))))(((....)))...... ( -24.90, z-score = -3.64, R) >droSec1.super_35 116230 91 + 471130 GUUUACUGCACUUUAAUAAAAUUGUUGGGUCACG--CGUUUAGUUUAAAACUGCACUGGUUUUAAUCACUGUGUAGUGCCCCUUGGCACGCAG .....((((......................(((--((......((((((((.....))))))))....))))).(((((....))))))))) ( -21.70, z-score = -0.46, R) >consensus _____________U_AUAAAAUUCUGGGGUCACA__CGUUAAGUUCAAGACUGCACUGGUUUUAAUCGCAGUGUAGUGCCCCUUGGCACGCAG ...................................................(((((((..........)))))))(((((....))))).... (-11.57 = -12.58 + 1.00)

| Location | 8,266,346 – 8,266,439 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.53529 |
| G+C content | 0.46451 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -10.74 |
| Energy contribution | -11.55 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
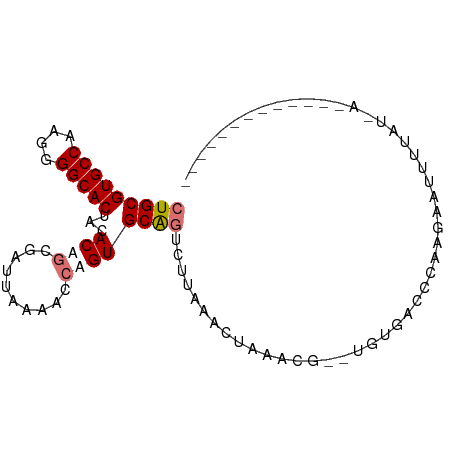
>dm3.chrX 8266346 93 - 22422827 CUGCGUGCCAAGGGGCACUACACAGCGCUUAAAACCAGUGCAGUCUUAAACUAAGCGUAUGUGACCCCCGAACUUUAUGAAAGUGCAUUAAAC .((((((((....)))))..(((((((((((.....((......)).....))))))).)))).....................)))...... ( -22.80, z-score = -0.56, R) >droEre2.scaffold_4690 10631797 79 - 18748788 CUGCGUGCCAAGGGGCACUACACUGGGAUUAAAACCAGUGCGCUCUAGAACUUCACACAUCUGACCCAA-AAGGUUAUCA------------- ..(((((((....)))))..((((((........)))))).))..................(((((...-..)))))...------------- ( -23.20, z-score = -1.99, R) >droYak2.chrX 8723158 76 + 21770863 CUGCGUGCCAAGGGGCACUACACUGCGAUUAAAGCGAGUGCAGUCCUGAAACUAAUG--UUUAGAUUCAGGGUAUCAU--------------- (((((((((....)))))...(((((.......)).)))))))(((((((.(((...--..))).)))))))......--------------- ( -26.00, z-score = -2.71, R) >droSec1.super_35 116230 91 - 471130 CUGCGUGCCAAGGGGCACUACACAGUGAUUAAAACCAGUGCAGUUUUAAACUAAACG--CGUGACCCAACAAUUUUAUUAAAGUGCAGUAAAC (((((((((....)))))..(((.((..(((((((.......))))))).....)).--.))).....................))))..... ( -19.30, z-score = -0.11, R) >consensus CUGCGUGCCAAGGGGCACUACACAGCGAUUAAAACCAGUGCAGUCUUAAACUAAACG__UGUGACCCAAGAAUUUUAU_A_____________ (((((((((....)))))...((((..........)))))))).................................................. (-10.74 = -11.55 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:39 2011