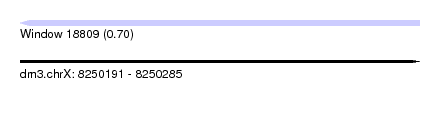
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,250,191 – 8,250,285 |
| Length | 94 |
| Max. P | 0.702657 |

| Location | 8,250,191 – 8,250,285 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.99 |
| Shannon entropy | 0.63252 |
| G+C content | 0.42714 |
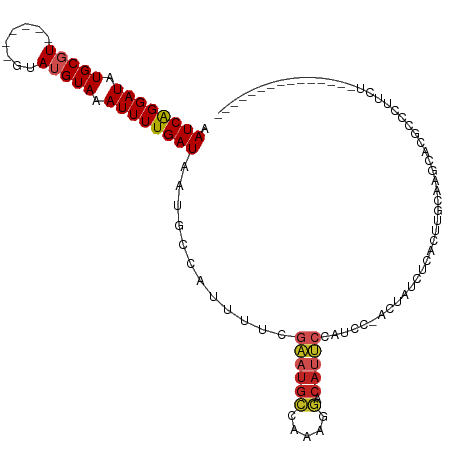
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -9.99 |
| Energy contribution | -10.45 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
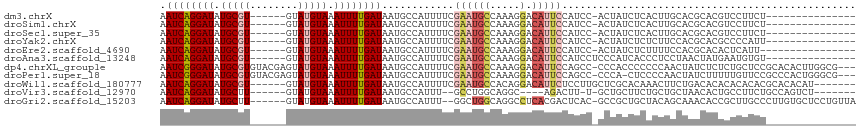
>dm3.chrX 8250191 94 - 22422827 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC-ACUAUCUCACUUGCACGCACGUCCUUCU--------------- ....((((..(((((------(((.((.......((((.((........(((((((...)).)))))....)-).))))..)).))))))))..))))...--------------- ( -23.70, z-score = -2.90, R) >droSim1.chrX 6575714 94 - 17042790 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC-ACUAUCUCACUUGCACGCACGUCCUUCU--------------- ....((((..(((((------(((.((.......((((.((........(((((((...)).)))))....)-).))))..)).))))))))..))))...--------------- ( -23.70, z-score = -2.90, R) >droSec1.super_35 100830 94 - 471130 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC-ACUAUCUCACUUGCACGCACGUCCUUCU--------------- ....((((..(((((------(((.((.......((((.((........(((((((...)).)))))....)-).))))..)).))))))))..))))...--------------- ( -23.70, z-score = -2.90, R) >droYak2.chrX 8707084 94 + 21770863 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC-ACUAUCUCUCUUCCACGCACGCCCCAUU--------------- .....((....((((------(..((........((((.((........(((((((...)).)))))....)-).)))).......))..))))).))...--------------- ( -16.86, z-score = -0.88, R) >droEre2.scaffold_4690 10617121 93 - 18748788 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC-ACUAUCUCUUUUCCACGCACACUCAUU---------------- .....(..(.(((((------(............((...........))(((((((...)).))))).....-.............)))))).)..)...---------------- ( -13.00, z-score = 0.20, R) >droAna3.scaffold_13248 653811 95 - 4840945 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCCUCCCAUCACCCUCCUAACUAUGAAUGUGU--------------- .((((((((.((((.------...)))).))))))))............(((((((...)).)))))...........(((..((........))..))).--------------- ( -13.00, z-score = 0.68, R) >dp4.chrXL_group1e 10647712 112 + 12523060 AAUCGGGAUAUGCGUGUACGAGUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAGCC-CCCACCCCCCCAACUAUCUCUCUGCUCCGCACACUUGGCG--- .((((((((.((((((......)))))).))))))))...((((.....(((((((...)).))))).....-......................(((...)))....)))).--- ( -20.30, z-score = 0.39, R) >droPer1.super_18 1226937 111 + 1952607 AAUCGGGAUAUGCGUGUACGAGUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAGCC-CCCA-CUCCCCAACUAUCUUUUUGUUCCGCCCACUGGGCG--- ....(((((((((........))))))......................(((((((...)).))))).....-))).-......................(((((...)))))--- ( -21.40, z-score = 0.47, R) >droWil1.scaffold_180777 4531961 103 + 4753960 AAUCAGGAUAUGCGU------GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCACAGGACAUUCUCCUUGCUCGCACAAACUUCUGACACACACACACGCACACAU------- ..........(((((------((.(((....(((((...........)))))(((((.((((.....))))))...)))..............))).))))))).....------- ( -18.20, z-score = -0.29, R) >droVir3.scaffold_12970 2277280 95 - 11907090 AAUCAGGAUAUGCUU------GUAUGUAAAUUUUGAUAAUGCCAUUU--GCCUGGCAGGC----AGACUU-U-GCUGCUUCUGCUGCUAACACUGCCUUCUGCCAGUCU------- .((((((((.(((..------....))).))))))))..........--..(((((((((----((....-.-((.((....)).)).....)))))...))))))...------- ( -30.10, z-score = -2.22, R) >droGri2.scaffold_15203 11352197 107 + 11997470 AAUCAGGAUAUGCUU------GUAUGUAAAUUUUGAUAAUGCCAUUU--GGCUGGCAGGCCUCACGACUCAC-GCCGCUGCUACAGCAAACACCGCUUGCCCUUGUGCUCCUGUUA .((((((((.(((..------....))).))))))))...((((...--(((.(((.(((............-)))((((...)))).......))).)))..)).))........ ( -27.10, z-score = 0.01, R) >consensus AAUCAGGAUAUGCGU______GUAUGUAAAUUUUGAUAAUGCCAUUUUCGAAUGCCAAAGGACAUUCCAUCC_ACUAUCUCACUUGCAAGCACGCCCUUCU_______________ .((((((((.(((((........))))).))))))))............((((((.....).)))))................................................. ( -9.99 = -10.45 + 0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:37 2011