| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,227,163 – 8,227,271 |
| Length | 108 |
| Max. P | 0.500000 |

| Location | 8,227,163 – 8,227,271 |
|---|---|
| Length | 108 |
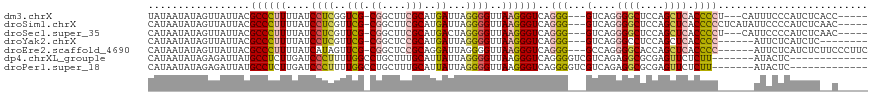
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Shannon entropy | 0.40175 |
| G+C content | 0.50251 |
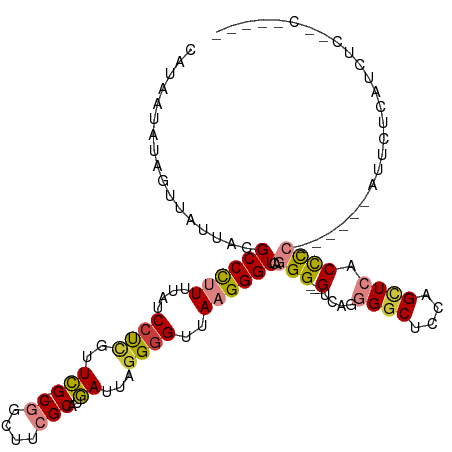
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -16.29 |
| Energy contribution | -15.16 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 8227163 108 - 22422827 UAUAAUAUAGUUAUUACGCCCUUUUAUCCUCGGUCG-CGGCUUCGCAUGAUUAGGGGUUAAGGGUCAGGG---GUCAGGGGCUCCAGCUCACCCCU---CAUUUCCCAUCUCACC----- ..(((((....))))).((((((..(((((..((((-((....)))..)))..))))).)))))).((((---((...((((....))))))))))---................----- ( -34.40, z-score = -1.77, R) >droSim1.chrX 6556439 111 - 17042790 CAUAAUAUAGUUAUUACGCCCUUUUAUCCUCGUUCG-CGGCUUCGCAUGAUUAGGGGUUAAGGGUCAGGG---GUCAGGGGCUCCAGCUCACCCCCUCAUAUUCCCCAUCUCAAC----- ..(((((....))))).((((((...((((.(((.(-((....)))..))).))))...))))))..(((---(..(((((...........)))))......))))........----- ( -30.70, z-score = -0.86, R) >droSec1.super_35 78911 108 - 471130 CAUAAUAUAGUUAUUACGCCCUUUUAUCCUCGUUCG-CGGCUUCGCAUGACUAGGGGUUAAGGGUCAGGG---GUCAGGGGCUCCAGCUCACCCCU---CAUUCCCCAUCUCAAC----- ..(((((....))))).((((((...((((.(((.(-((....)))..))).))))...))))))..(((---(..(((((..........)))))---....))))........----- ( -33.90, z-score = -2.12, R) >droYak2.chrX 8683513 102 + 21770863 CAUAAUAUAGUUAUUACGCCCUUUUAUCCUCGUUCG-CGGCUCCGCAUGAUUAGGGGUUAAGGGUCAGGG---GUCAGGGCCUCCAGCUCACCCC------AUUCUCAUCUC-------- ..(((((....))))).((((((...((((.(((.(-((....)))..))).))))...))))))..(((---((..((((.....)))))))))------...........-------- ( -31.90, z-score = -2.18, R) >droEre2.scaffold_4690 10595134 110 - 18748788 CAUAAUAUAGUUAUUACGCCCUUUUAUCAUAGUUCG-CGGCUCCGCAGGAUUAGGGGUUAAGGGUCAGGG---GCCAGGGGCACCAGCUCACCCC------AUUCUCAUCUCUUCCCUUC ..(((((....))))).((((((..(((.(((((((-((....))).))))))..))).))))))..(((---(....((((....)))).))))------................... ( -31.70, z-score = -1.10, R) >dp4.chrXL_group1e 10622215 100 + 12523060 CAUAAUAUAGAGAUUAUGCCUCUUGAUCCCUUUUGGCCUGCUUUGCAUUAUUAGGGGUUAAGGGUCAGGGGUCGUCAGAGGCGCGAGUUCUCUU-------AUACUC------------- .....(((((((((..(((((((((((((((...((((((((((.........)))))...)))))))))))))..)))))))..)..)))).)-------)))...------------- ( -33.50, z-score = -2.19, R) >droPer1.super_18 1201000 100 + 1952607 CAUAAUAUAGAGAUUAUGCCUCUUGAUCCCUUUUGGCCUGCUUUGCAUUAUUAGGGGUUAAGGGUCAGGGGUCGUCAGAGGCGCGAGUUCUCUU-------AUACUC------------- .....(((((((((..(((((((((((((((...((((((((((.........)))))...)))))))))))))..)))))))..)..)))).)-------)))...------------- ( -33.50, z-score = -2.19, R) >consensus CAUAAUAUAGUUAUUACGCCCUUUUAUCCUCGUUCG_CGGCUUCGCAUGAUUAGGGGUUAAGGGUCAGGG___GUCAGGGGCUCCAGCUCACCCC______AUUCUCAUCUC__C_____ .................((((((....((((..(((...((...)).)))...))))..))))))..(((.......(((((....))...)))..........)))............. (-16.29 = -15.16 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:32 2011