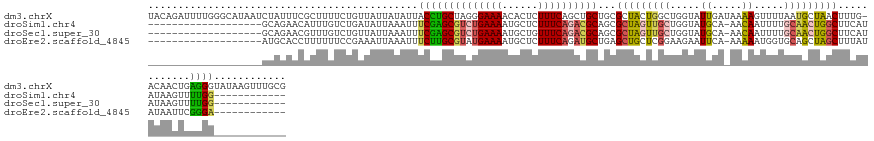
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,189,458 – 8,189,600 |
| Length | 142 |
| Max. P | 0.976835 |

| Location | 8,189,458 – 8,189,600 |
|---|---|
| Length | 142 |
| Sequences | 4 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 61.42 |
| Shannon entropy | 0.55889 |
| G+C content | 0.38610 |
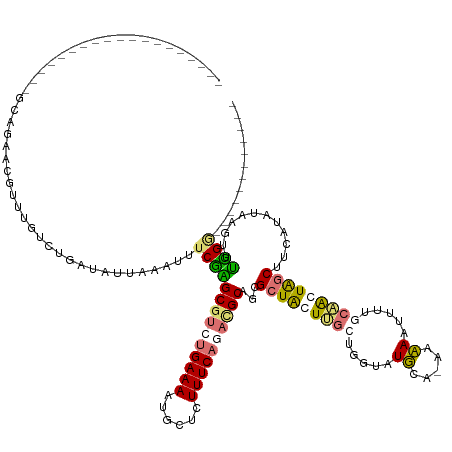
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -13.41 |
| Energy contribution | -16.10 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
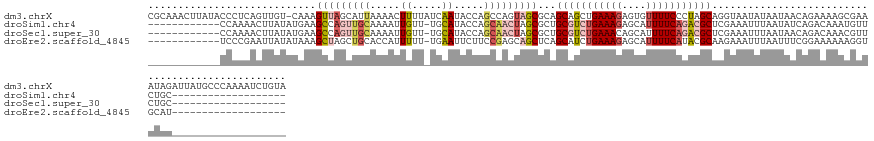
>dm3.chrX 8189458 142 + 22422827 CGCAAACUUAUACCCUCAGUUGU-CAAAGUUAGCAUUAAAACUUUUAUCAAUACCAGCCAGUAGCGCAGCAGCUGAAAGAGUGUUUUCCCUAGCAGGUAAUAUAAUAACAGAAAAGCGAAAUAGAUUAUGCCCAAAAUCUGUA (((............((((((((-.((((((........))))))...........((.....))...))))))))....(((((..((......)).)))))............)))..(((((((........))))))). ( -23.10, z-score = 0.02, R) >droSim1.chr4 577828 111 + 949497 ------------CCAAAACUUAUAUGAAGCCAGUUGCAAAAUUGUU-UGCAUACCAGCAACUAGCGCUGCGUCUGAAAGAGCAUUUUCAGACGCUCGAAAUUUAAUAUCAGACAAAUGUUCUGC------------------- ------------.((.(((.....(((.((.((((((.....((..-..)).....)))))).))...(((((((((((....))))))))))).............))).......))).)).------------------- ( -28.00, z-score = -1.96, R) >droSec1.super_30 461112 111 + 664411 ------------CCAAAACUUAUAUGAAGCCAGUUGCAAAAUUGUU-UGCAUACCAGCAACUAGCGCUGCGUCUGAAACAGCAUUUUCAGACGCUCGAAAUUUAAUAACAGACAAACGUUCUGC------------------- ------------............(((.((.((((((.....((..-..)).....)))))).))...((((((((((......)))))))))))))...........((((.......)))).------------------- ( -27.90, z-score = -2.30, R) >droEre2.scaffold_4845 20863497 111 - 22589142 ------------UCCCGAAUUAUAUAAAGCUAGCUGCACCAUUUUU-UGAAUUCUUCCGAGCAGCUCAGCAUCUGAAAGAGCAUUUUCAUACGCAAGAAAUUUAAUUUCGGAAAAAAGGUGCAU------------------- ------------..((............(((((((((..((.....-))...((....)))))))).))).(((((((......((((........)))).....))))))).....)).....------------------- ( -22.60, z-score = -0.75, R) >consensus ____________CCAAAACUUAUAUAAAGCCAGCUGCAAAAUUGUU_UGAAUACCAGCAACUAGCGCAGCAUCUGAAAGAGCAUUUUCAGACGCACGAAAUUUAAUAACAGAAAAACGUUCUAC___________________ ............................(((((((((.....((.....)).....)))))))))...(((((((((((....)))))))))))................................................. (-13.41 = -16.10 + 2.69)
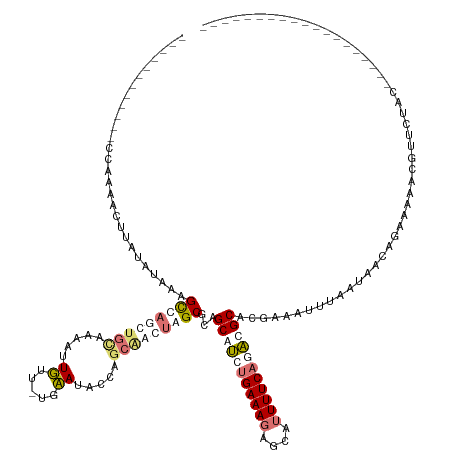
| Location | 8,189,458 – 8,189,600 |
|---|---|
| Length | 142 |
| Sequences | 4 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 61.42 |
| Shannon entropy | 0.55889 |
| G+C content | 0.38610 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8189458 142 - 22422827 UACAGAUUUUGGGCAUAAUCUAUUUCGCUUUUCUGUUAUUAUAUUACCUGCUAGGGAAAACACUCUUUCAGCUGCUGCGCUACUGGCUGGUAUUGAUAAAAGUUUUAAUGCUAACUUUG-ACAACUGAGGGUAUAAGUUUGCG .(((((....((((............)))).)))))(((((...((((.(((((.((((......))))(((......))).))))).)))).)))))...........((.(((((..-((........))..))))).)). ( -29.20, z-score = 0.19, R) >droSim1.chr4 577828 111 - 949497 -------------------GCAGAACAUUUGUCUGAUAUUAAAUUUCGAGCGUCUGAAAAUGCUCUUUCAGACGCAGCGCUAGUUGCUGGUAUGCA-AACAAUUUUGCAACUGGCUUCAUAUAAGUUUUGG------------ -------------------.((((((..((((.(((.............((((((((((......))))))))))...(((((((((.....((..-..)).....))))))))).))).)))))))))).------------ ( -36.70, z-score = -3.63, R) >droSec1.super_30 461112 111 - 664411 -------------------GCAGAACGUUUGUCUGUUAUUAAAUUUCGAGCGUCUGAAAAUGCUGUUUCAGACGCAGCGCUAGUUGCUGGUAUGCA-AACAAUUUUGCAACUGGCUUCAUAUAAGUUUUGG------------ -------------------.((((((.......................((((((((((......))))))))))...(((((((((.....((..-..)).....))))))))).........)))))).------------ ( -35.30, z-score = -2.66, R) >droEre2.scaffold_4845 20863497 111 + 22589142 -------------------AUGCACCUUUUUUCCGAAAUUAAAUUUCUUGCGUAUGAAAAUGCUCUUUCAGAUGCUGAGCUGCUCGGAAGAAUUCA-AAAAAUGGUGCAGCUAGCUUUAUAUAAUUCGGGA------------ -------------------...........(((((((............((((.(((((......))))).))))..((((((((....))..(((-.....))).))))))............)))))))------------ ( -25.90, z-score = -0.77, R) >consensus ___________________GCAGAACGUUUGUCUGAUAUUAAAUUUCGAGCGUCUGAAAAUGCUCUUUCAGACGCAGCGCUACUUGCUGGUAUGCA_AAAAAUUUUGCAACUAGCUUCAUAUAAGUUUGGG____________ ..............................................(((((((((((((......))))))))))...(((((((((.....((.....)).....)))))))))............)))............. (-17.66 = -17.98 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:30 2011