
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,168,084 – 8,168,142 |
| Length | 58 |
| Max. P | 0.999231 |

| Location | 8,168,084 – 8,168,142 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 59.39 |
| Shannon entropy | 0.65872 |
| G+C content | 0.37146 |
| Mean single sequence MFE | -13.15 |
| Consensus MFE | -8.06 |
| Energy contribution | -7.67 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
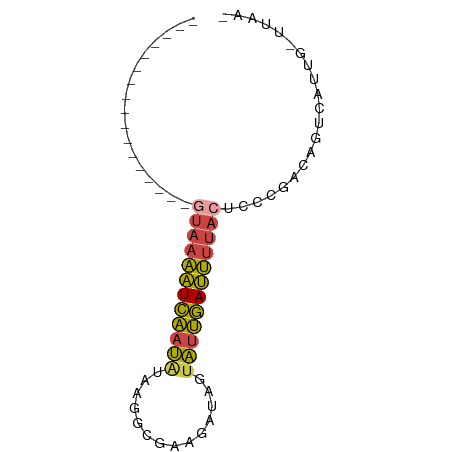
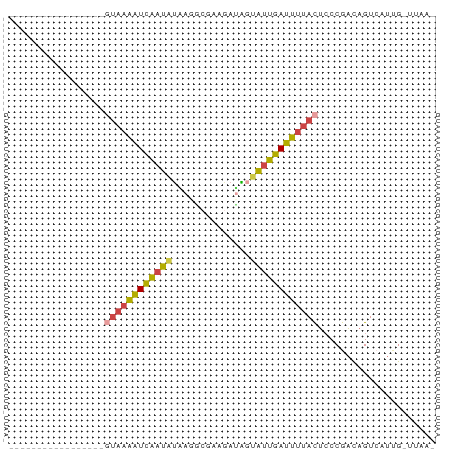
>dm3.chrX 8168084 58 + 22422827 -----------------GUAAAAUCAAUAUAAGGCGAAGAUAGUAUUGAUUUUACUCCCGACAGUCAUUGUUUAA- -----------------(((((((((((((............)))))))))))))....................- ( -11.50, z-score = -2.43, R) >droEre2.scaffold_4690 10533147 57 + 18748788 -----------------GUAAAAUCAAUAUUAGACCAGAAAGCUAUUGAUUUUACUCCCGACAGUCUUAG-UUAA- -----------------((((((((((((..............))))))))))))...............-....- ( -9.74, z-score = -2.54, R) >droYak2.chrX 8623897 59 - 21770863 -----------------UCAAAAUCAAUAUUAGACCAAGAUAGUAUUGAUUUUACUCCCGACAGUCGUAGGUUAAG -----------------..(((((((((((((........)))))))))))))(((..((.....))..))).... ( -13.50, z-score = -3.82, R) >droSec1.super_35 19679 53 + 471130 -----------------GUAAAAUCAAUGUAAAGCGAAGAUAGUAUUGAUUUUACUCCCGACAGUGCUUG------ -----------------((((((((((((..............))))))))))))...............------ ( -9.54, z-score = -1.91, R) >droSim1.chrX 6503263 53 + 17042790 -----------------GUAAAAUCAAUGUAAGGCGAAGAUAGUAUUGAUUUUACUCCCGACAGUGUUUG------ -----------------((((((((((((..(........)..))))))))))))...............------ ( -10.20, z-score = -1.77, R) >droWil1.scaffold_180777 4443538 75 - 4753960 AUAGACCCAAUCUGGCCCUGCUGUGGGCGAUAGCAAAGGGGUCAGGCCACAUAAUGACCGCUAGUCAUCGCCUAA- ...(((((...(((((((......))))..))).....)))))((((......(((((.....))))).))))..- ( -24.40, z-score = -0.62, R) >consensus _________________GUAAAAUCAAUAUAAGGCGAAGAUAGUAUUGAUUUUACUCCCGACAGUCAUUG_UUAA_ .................((((((((((((..............))))))))))))..................... ( -8.06 = -7.67 + -0.38)

| Location | 8,168,084 – 8,168,142 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 59.39 |
| Shannon entropy | 0.65872 |
| G+C content | 0.37146 |
| Mean single sequence MFE | -11.41 |
| Consensus MFE | -6.93 |
| Energy contribution | -6.24 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
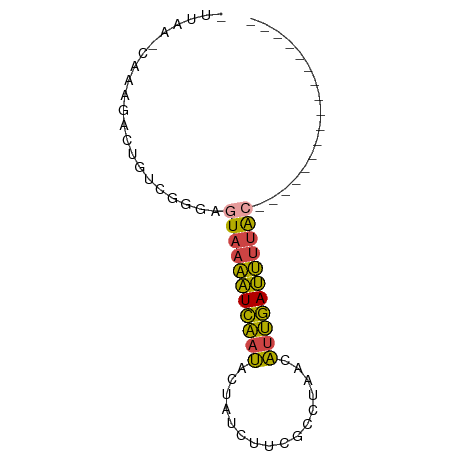
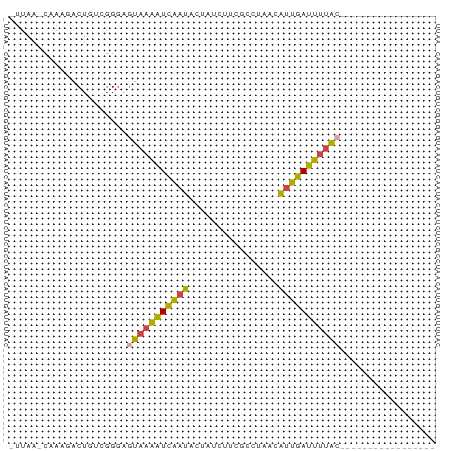
>dm3.chrX 8168084 58 - 22422827 -UUAAACAAUGACUGUCGGGAGUAAAAUCAAUACUAUCUUCGCCUUAUAUUGAUUUUAC----------------- -....................((((((((((((.((.........))))))))))))))----------------- ( -8.60, z-score = -1.34, R) >droEre2.scaffold_4690 10533147 57 - 18748788 -UUAA-CUAAGACUGUCGGGAGUAAAAUCAAUAGCUUUCUGGUCUAAUAUUGAUUUUAC----------------- -....-...............(((((((((((((((....)))....))))))))))))----------------- ( -10.50, z-score = -1.50, R) >droYak2.chrX 8623897 59 + 21770863 CUUAACCUACGACUGUCGGGAGUAAAAUCAAUACUAUCUUGGUCUAAUAUUGAUUUUGA----------------- .....(((((....)).)))..(((((((((((.((........)).))))))))))).----------------- ( -9.30, z-score = -0.69, R) >droSec1.super_35 19679 53 - 471130 ------CAAGCACUGUCGGGAGUAAAAUCAAUACUAUCUUCGCUUUACAUUGAUUUUAC----------------- ------...............(((((((((((................)))))))))))----------------- ( -7.29, z-score = -1.18, R) >droSim1.chrX 6503263 53 - 17042790 ------CAAACACUGUCGGGAGUAAAAUCAAUACUAUCUUCGCCUUACAUUGAUUUUAC----------------- ------...............(((((((((((................)))))))))))----------------- ( -7.29, z-score = -1.36, R) >droWil1.scaffold_180777 4443538 75 + 4753960 -UUAGGCGAUGACUAGCGGUCAUUAUGUGGCCUGACCCCUUUGCUAUCGCCCACAGCAGGGCCAGAUUGGGUCUAU -..((((((((((.....)))))).....))))(((((.((((.....((((......))))))))..)))))... ( -25.50, z-score = -0.82, R) >consensus _UUAA_CAAAGACUGUCGGGAGUAAAAUCAAUACUAUCUUCGCCUAACAUUGAUUUUAC_________________ .....................(((((((((((................)))))))))))................. ( -6.93 = -6.24 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:28 2011