| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,113,309 – 8,113,387 |
| Length | 78 |
| Max. P | 0.936881 |

| Location | 8,113,309 – 8,113,387 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.55577 |
| G+C content | 0.42238 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -7.82 |
| Energy contribution | -9.74 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936881 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
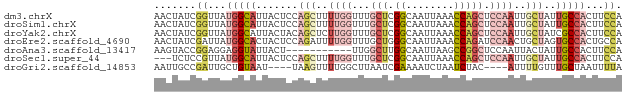
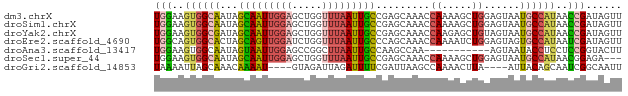
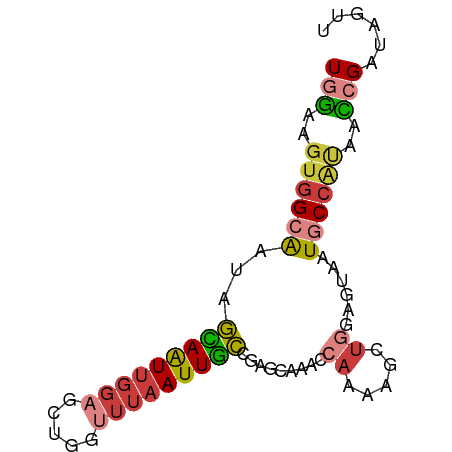
>dm3.chrX 8113309 78 + 22422827 AACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCA .......((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)). ( -21.60, z-score = -2.96, R) >droSim1.chrX 6468723 78 + 17042790 AACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCA .......((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)). ( -21.60, z-score = -2.96, R) >droYak2.chrX 8569615 78 - 21770863 AACUAUCGGUUAUGGCAUUACUACAGCUCUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUCGCCACUUCCA .......((...((((........(((..((((...(((.((........))))).))))..)))...))))...)). ( -18.50, z-score = -2.11, R) >droEre2.scaffold_4690 10480788 78 + 18748788 AACUAUCGAUUAUGGCACUACUCCAGAUUUUGGUUUGCUGGGCAAUUAAACCAGAUCCAACUGCUAGUGCCACUGCCA ............((((((((...(((...((((....((((.........))))..))))))).))))))))...... ( -22.50, z-score = -2.36, R) >droAna3.scaffold_13417 461671 67 + 6960332 AAGUACCGGAGGAGGUAUUACU-----------UUGGCUUGGCAAUUAAGCCGGCUCCAAUUACUAUUGCCACUUCCA .......(((((.((((.((..-----------((((.(((((......)))))..))))....)).)))).))))). ( -21.20, z-score = -2.04, R) >droSec1.super_44 204465 75 + 251150 ---UCUCCGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCA ---.........(((((.......(((..((((...(((.((........))))).))))..)))..)))))...... ( -18.60, z-score = -2.48, R) >droGri2.scaffold_14853 1979253 70 - 10151454 AAUUGCCGAUUGCUGUAAU----UAAGUUUUGGCUUAAUCGAAAAUCUAAUCUAC----AUUUUGUUUGCUAAUUUUA ....(((((..(((.....----..))).))))).....................----................... ( -8.00, z-score = 0.06, R) >consensus AACUAUCGGUUAUGGCAUUACUCCAGCUUUUGGUUUGCUCGGCAAUUAAACCAGCUCCAAUUGCUAUUGCCACUUCCA .......((...(((((.......(((..((((...(((.((........))))).))))..)))..)))))...)). ( -7.82 = -9.74 + 1.92)
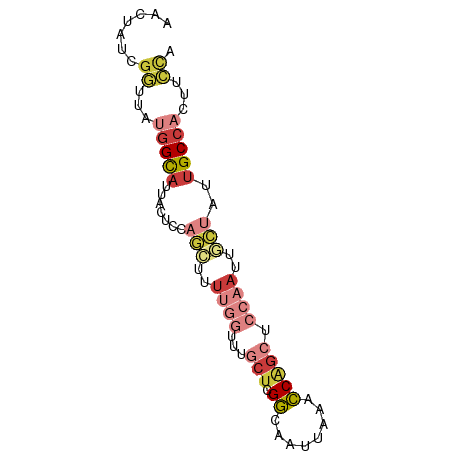
| Location | 8,113,309 – 8,113,387 |
|---|---|
| Length | 78 |
| Sequences | 7 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.55577 |
| G+C content | 0.42238 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -7.90 |
| Energy contribution | -8.47 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712786 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 8113309 78 - 22422827 UGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUU .((..((((((.((((..((((.((((((......))).)))...))))..))))......))))))..))....... ( -23.30, z-score = -2.34, R) >droSim1.chrX 6468723 78 - 17042790 UGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUU .((..((((((.((((..((((.((((((......))).)))...))))..))))......))))))..))....... ( -23.30, z-score = -2.34, R) >droYak2.chrX 8569615 78 + 21770863 UGGAAGUGGCGAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAGAGCUGUAGUAAUGCCAUAACCGAUAGUU .((..(((((((((((..((((.((((((......))).)))...))))..))))).....))))))..))....... ( -25.20, z-score = -3.19, R) >droEre2.scaffold_4690 10480788 78 - 18748788 UGGCAGUGGCACUAGCAGUUGGAUCUGGUUUAAUUGCCCAGCAAACCAAAAUCUGGAGUAGUGCCAUAAUCGAUAGUU .((..(((((((((((((((((((...))))))))))((((...........))))..)))))))))..))....... ( -24.70, z-score = -2.07, R) >droAna3.scaffold_13417 461671 67 - 6960332 UGGAAGUGGCAAUAGUAAUUGGAGCCGGCUUAAUUGCCAAGCCAA-----------AGUAAUACCUCCUCCGGUACUU (((...((((((((((...((....)))))..)))))))..))).-----------.....((((......))))... ( -16.20, z-score = -0.24, R) >droSec1.super_44 204465 75 - 251150 UGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACGGAGA--- .....((((((.((((..((((.((((((......))).)))...))))..))))......))))))........--- ( -20.90, z-score = -1.70, R) >droGri2.scaffold_14853 1979253 70 + 10151454 UAAAAUUAGCAAACAAAAU----GUAGAUUAGAUUUUCGAUUAAGCCAAAACUUA----AUUACAGCAAUCGGCAAUU ........((........(----((.((........))(((((((......))))----)))))).......)).... ( -7.06, z-score = -0.46, R) >consensus UGGAAGUGGCAAUAGCAAUUGGAGCUGGUUUAAUUGCCGAGCAAACCAAAAGCUGGAGUAAUGCCAUAACCGAUAGUU (((..((((((...(((((((((.....))))))))).........((.....))......))))))..)))...... ( -7.90 = -8.47 + 0.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:20 2011