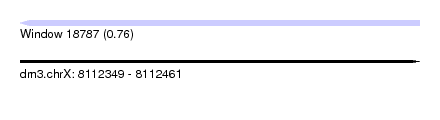
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,112,349 – 8,112,461 |
| Length | 112 |
| Max. P | 0.758379 |

| Location | 8,112,349 – 8,112,461 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 64.90 |
| Shannon entropy | 0.66299 |
| G+C content | 0.36110 |
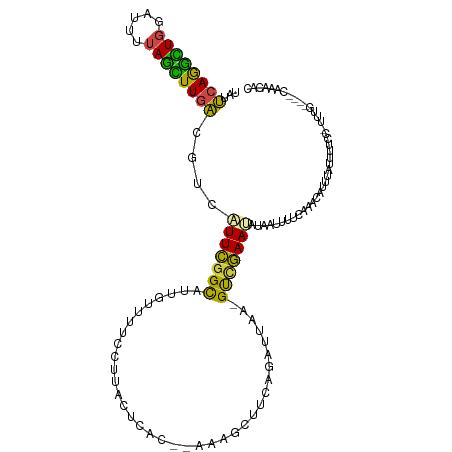
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -8.55 |
| Energy contribution | -7.34 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.98 |
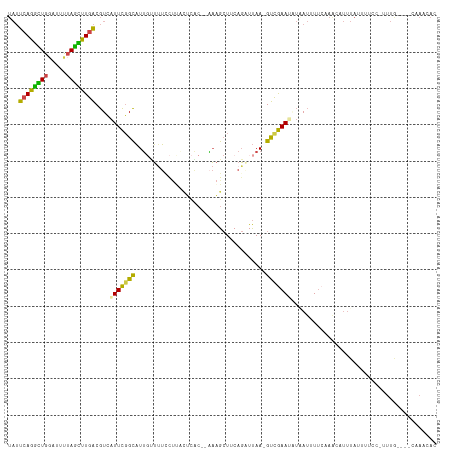
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
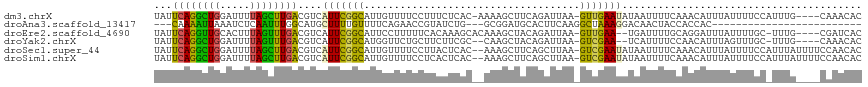
>dm3.chrX 8112349 112 - 22422827 UAUUCAGGCUGGAUUUUAGCUUGACGUCAUUCGGCAUUGUUUUCCUUUCUCAC-AAAAGCUUCAGAUUAA-GUUGAAUAUAAUUUUCAAACAUUUAUUUUCCAUUUG----CAAACAC (((((((.((.(((((.(((((...(((....))).((((...........))-)))))))..))))).)-))))))))............................----....... ( -14.30, z-score = 0.40, R) >droAna3.scaffold_13417 460743 87 - 6960332 ---CAAAAUUAAAUCUCAAUUUGGCAUGCUUUUGUUUUCAGAACCGUAUCUG---GCGGAUGCACUUCAAGGCUAAAGGACAACUACCACCAC------------------------- ---................((((((..(.(((((....))))).)(((((..---...)))))........))))))((.......)).....------------------------- ( -13.00, z-score = 0.58, R) >droEre2.scaffold_4690 10479903 110 - 18748788 UAUUCAGGUUGCACUUUAGUUUGACGUCAUUCGGCAUUCCUUUUUCACAAAGCACAAAGCUACAGAUUAA-GUUGAA--UGAUUUUGCAGGAUUUAUUUUGC-UUUG----CGAUCAC ......(((((((.........((.((((((((((.....((..((....(((.....)))...))..))-))))))--)))).))((((((....))))))-..))----))))).. ( -23.10, z-score = -1.08, R) >droYak2.chrX 8568628 108 + 21770863 UAUUCAGGCUGGAUUUUAGUUUGACGUCAUUCGGCAUGGUUCUGCUUCUUCGC--CAAGCUACAGAUUAA-GUCGAA--UCAUUUUCCAACAUUUAGUUUGC-UUUG----CAAACAC .((((.((((.((((((((((((.((......((((......))))....)).--))))))).))))).)-))))))--)................((((((-...)----))))).. ( -24.80, z-score = -1.62, R) >droSec1.super_44 203480 115 - 251150 UAUUCAGGCUGGAUUUUAGCUUGACGUCAUUCGGCAUUGUUUUCCUUACUCAC--AAAGCUUCAGCUUAA-GUCGAAUAUAAUUUUCAAACAUUUAUUUUCCAUUUAUUUUCCAACAC ...((((((((.....))))))))....(((((((...((.......))....--.((((....))))..-)))))))........................................ ( -18.20, z-score = -2.07, R) >droSim1.chrX 6467691 115 - 17042790 UAUUCAGGCUGGAUUUUAGCUUGACGUCAUUCGGCAUUGUUUUCCUCACUCAC--AAAGCUUCAGCUUAA-GUCGAAUAUAAUUUUCAAACAUUUAUUUUCCAUUUAUUUUCCAACAC ...((((((((.....))))))))....(((((((..(((...........))--)((((....))))..-)))))))........................................ ( -18.20, z-score = -2.11, R) >consensus UAUUCAGGCUGGAUUUUAGCUUGACGUCAUUCGGCAUUGUUUUCCUUACUCAC__AAAGCUUCAGAUUAA_GUCGAAUAUAAUUUUCAAACAUUUAUUUUCC_UUUG____CAAACAC ...((((((((.....))))))))....(((((((....................................)))))))........................................ ( -8.55 = -7.34 + -1.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:18 2011