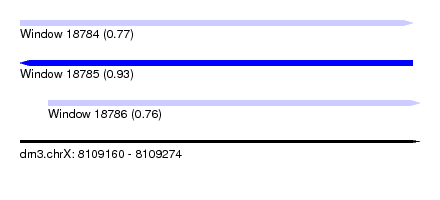
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,109,160 – 8,109,274 |
| Length | 114 |
| Max. P | 0.931595 |

| Location | 8,109,160 – 8,109,272 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 64.16 |
| Shannon entropy | 0.71723 |
| G+C content | 0.45881 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -11.76 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
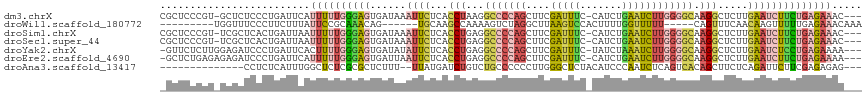
>dm3.chrX 8109160 112 + 22422827 CGCUCCCGU-GCUCUCCCUGAUUCAUUUUUGGGAGUGAUAAAUUCUCACCUAAGGCCCCAGCUUCGAUUUC-CAUCUGAAUCUUGGGGCAAGGCUCUUGAAUCUUCUGAGAAAC--- .((......-))((((...((((((....(((((((.....)))))))(((...(((((((.((((((...-.))).)))..))))))).)))....))))))....))))...--- ( -34.00, z-score = -1.68, R) >droWil1.scaffold_180772 2006559 97 + 8906247 ---------UGGUUUCCCUUCUUUAUUCCGCAAACAG------UGCAAGCCAAAAGUCUAGGCUUAAGUCCACUUUUGGUUUUU-----CAGUUUCAACAAGUUUUUGAGAAACAAA ---------..(((((.............(((.....------)))(((((((((((...(((....))).)))))))))))..-----....................)))))... ( -19.70, z-score = -1.12, R) >droSim1.chrX 6464609 112 + 17042790 CGCUCCCGU-UCGCUCACUGAUUAAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUC-CAUCUGAAUCUUGGGGCAAGGCUCUUGAAUCUUCUGAGAAAC--- ((((((((.-...................)))))))).....(((((((((...(((((((.((((((...-.))).)))..))))))).))).....((....))))))))..--- ( -33.25, z-score = -1.47, R) >droSec1.super_44 200368 112 + 251150 CGCUCCCGU-UCGCUCACUGAUUAAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUC-CAUCUGAAUCUUGGGGCAAGGCUCUUGAAUCUUCUGAGAAAC--- ((((((((.-...................)))))))).....(((((((((...(((((((.((((((...-.))).)))..))))))).))).....((....))))))))..--- ( -33.25, z-score = -1.47, R) >droYak2.chrX 8565324 112 - 21770863 -GUUCUCUUGGAGAUCCCUGAUUCACUUUUGGGAGUGAUAUAUUCUCACCUGAGGCCCCAGCUUCGAUUUC-UAUCUAAAUCUUGGGGCAAGGCUCUUGAAUCUCCUGAGAAAA--- -.(((((..((((((.(..((.((((((....))))))..........(((...(((((((....(((((.-.....)))))))))))).))).))..).)))))).)))))..--- ( -37.10, z-score = -2.05, R) >droEre2.scaffold_4690 10476782 112 + 18748788 -GCUCUGAGAGAGAUCCCUGAUUCAUUUUUGGGAGUGAUUAAUUCUCACCUGAGGCCCCAGCUUCGAUUUC-CAUCUGAAUCUUGGGGCAAGGCUCUUGAAUCUUCUGAGAAAA--- -..(((.((((((....))((((((....(((((((.....)))))))(((...(((((((.((((((...-.))).)))..))))))).)))....)))))))))).)))...--- ( -34.30, z-score = -0.36, R) >droAna3.scaffold_13417 457175 98 + 6960332 --------------CCUCUCAUUUGGCUCUCGCGCUCUUU--UUAUGAUCUGUCUGCCCCCCUUGGGCUCUACAUCCCAAUCUCAGUCACAGCUUCUCAGAUUCUUCGAGAGAG--- --------------.(((((....(((......)))....--....((((((...((((.....))))................(((....)))...))))))....)))))..--- ( -19.50, z-score = 0.06, R) >consensus _GCUCCCGU_GCGCUCCCUGAUUCAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUC_CAUCUGAAUCUUGGGGCAAGGCUCUUGAAUCUUCUGAGAAAC___ .........................((((((((((......((((...(((...(((((((....(((((.......)))))))))))).))).....))))))))))))))..... (-11.76 = -13.04 + 1.28)

| Location | 8,109,160 – 8,109,272 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.16 |
| Shannon entropy | 0.71723 |
| G+C content | 0.45881 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -12.57 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8109160 112 - 22422827 ---GUUUCUCAGAAGAUUCAAGAGCCUUGCCCCAAGAUUCAGAUG-GAAAUCGAAGCUGGGGCCUUAGGUGAGAAUUUAUCACUCCCAAAAAUGAAUCAGGGAGAGC-ACGGGAGCG ---((((((..((((((((....((((.((((((...(((.(((.-...))))))..))))))...))))..)))))).)).(((((............)))))...-..)))))). ( -38.30, z-score = -2.29, R) >droWil1.scaffold_180772 2006559 97 - 8906247 UUUGUUUCUCAAAAACUUGUUGAAACUG-----AAAAACCAAAAGUGGACUUAAGCCUAGACUUUUGGCUUGCA------CUGUUUGCGGAAUAAAGAAGGGAAACCA--------- ((((((((((((.......)))).....-----...........(..(((.((((((.........))))))..------..)))..)))))))))...((....)).--------- ( -22.60, z-score = -1.20, R) >droSim1.chrX 6464609 112 - 17042790 ---GUUUCUCAGAAGAUUCAAGAGCCUUGCCCCAAGAUUCAGAUG-GAAAUCGAAGCUGGGGCCUCAGGUGAGAAUUUAUCACUCCCAAAAAUUAAUCAGUGAGCGA-ACGGGAGCG ---((((((....((((((....((((.((((((...(((.(((.-...))))))..))))))...))))..)))))).(((((..............)))))....-..)))))). ( -31.84, z-score = -0.94, R) >droSec1.super_44 200368 112 - 251150 ---GUUUCUCAGAAGAUUCAAGAGCCUUGCCCCAAGAUUCAGAUG-GAAAUCGAAGCUGGGGCCUCAGGUGAGAAUUUAUCACUCCCAAAAAUUAAUCAGUGAGCGA-ACGGGAGCG ---((((((....((((((....((((.((((((...(((.(((.-...))))))..))))))...))))..)))))).(((((..............)))))....-..)))))). ( -31.84, z-score = -0.94, R) >droYak2.chrX 8565324 112 + 21770863 ---UUUUCUCAGGAGAUUCAAGAGCCUUGCCCCAAGAUUUAGAUA-GAAAUCGAAGCUGGGGCCUCAGGUGAGAAUAUAUCACUCCCAAAAGUGAAUCAGGGAUCUCCAAGAGAAC- ---..(((((.((((((((..((((((.((((((.(((((.....-.))))).....))))))...)))).........(((((......))))).))..))))))))..))))).- ( -39.30, z-score = -3.31, R) >droEre2.scaffold_4690 10476782 112 - 18748788 ---UUUUCUCAGAAGAUUCAAGAGCCUUGCCCCAAGAUUCAGAUG-GAAAUCGAAGCUGGGGCCUCAGGUGAGAAUUAAUCACUCCCAAAAAUGAAUCAGGGAUCUCUCUCAGAGC- ---....(((....((((((.(((....((((((...(((.(((.-...))))))..))))))))).(((((.......)))))........)))))).((((....)))).))).- ( -33.90, z-score = -0.90, R) >droAna3.scaffold_13417 457175 98 - 6960332 ---CUCUCUCGAAGAAUCUGAGAAGCUGUGACUGAGAUUGGGAUGUAGAGCCCAAGGGGGGCAGACAGAUCAUAA--AAAGAGCGCGAGAGCCAAAUGAGAGG-------------- ---..((((((..((.((((.....((((..((....(((((........))))).))..)))).))))))....--.....(.((....)))...)))))).-------------- ( -27.50, z-score = -1.08, R) >consensus ___GUUUCUCAGAAGAUUCAAGAGCCUUGCCCCAAGAUUCAGAUG_GAAAUCGAAGCUGGGGCCUCAGGUGAGAAUUUAUCACUCCCAAAAAUGAAUCAGGGAGCGC_ACGGGAGC_ ......((((...........(((....((((((...(((.(((.....))))))..))))))))).(((((.......)))))...............)))).............. (-12.57 = -13.30 + 0.73)
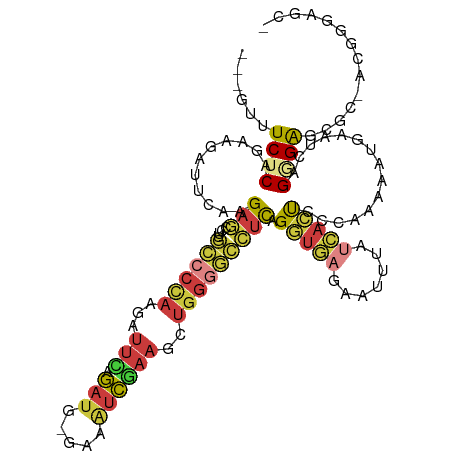
| Location | 8,109,168 – 8,109,274 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 62.68 |
| Shannon entropy | 0.70656 |
| G+C content | 0.44409 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.27 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.63 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
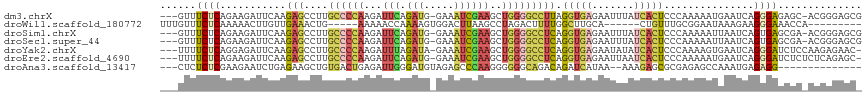
>dm3.chrX 8109168 106 + 22422827 UGCUCUCCCUGAUUCAUUUUUGGGAGUGAUAAAUUCUCACCUAAGGCCCCAGCUUCGAUUUCCAUCUGAAUCUUGGGGC--AAGGCUCUUGAAUCUUCUGAGAAACUG--------- ...((((...((((((....(((((((.....)))))))(((...(((((((.((((((....))).)))..)))))))--.)))....))))))....)))).....--------- ( -32.50, z-score = -1.87, R) >droWil1.scaffold_180772 2006559 105 + 8906247 ------------UGGUUUCCCUUCUUUAUUCCGCAAACAGUGCAAGCCAAAAGUCUAGGCUUAAGUCCACUUUUGGUUUUUCAGUUUCAACAAGUUUUUGAGAAACAAACUUUAAUG ------------..(((((.............(((.....)))(((((((((((...(((....))).)))))))))))......................)))))........... ( -19.70, z-score = -0.80, R) >droSim1.chrX 6464617 106 + 17042790 UUCGCUCACUGAUUAAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUCCAUCUGAAUCUUGGGGC--AAGGCUCUUGAAUCUUCUGAGAAACUG--------- .((((((.(..(.......)..)))))))....(((((((((...(((((((.((((((....))).)))..)))))))--.))).....((....))))))))....--------- ( -31.20, z-score = -1.62, R) >droSec1.super_44 200376 106 + 251150 UUCGCUCACUGAUUAAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUCCAUCUGAAUCUUGGGGC--AAGGCUCUUGAAUCUUCUGAGAAACUG--------- .((((((.(..(.......)..)))))))....(((((((((...(((((((.((((((....))).)))..)))))))--.))).....((....))))))))....--------- ( -31.20, z-score = -1.62, R) >droYak2.chrX 8565332 106 - 21770863 GGAGAUCCCUGAUUCACUUUUGGGAGUGAUAUAUUCUCACCUGAGGCCCCAGCUUCGAUUUCUAUCUAAAUCUUGGGGC--AAGGCUCUUGAAUCUCCUGAGAAAAUG--------- ((((((.(..((.((((((....))))))..........(((...(((((((....(((((......))))))))))))--.))).))..).))))))..........--------- ( -31.20, z-score = -0.86, R) >droEre2.scaffold_4690 10476790 106 + 18748788 AGAGAUCCCUGAUUCAUUUUUGGGAGUGAUUAAUUCUCACCUGAGGCCCCAGCUUCGAUUUCCAUCUGAAUCUUGGGGC--AAGGCUCUUGAAUCUUCUGAGAAAAUG--------- ..(((.....((((((....(((((((.....)))))))(((...(((((((.((((((....))).)))..)))))))--.)))....)))))).))).........--------- ( -30.10, z-score = -0.22, R) >droAna3.scaffold_13417 457183 92 + 6960332 ------------UUGGCUCUCGCGCUCUUUUUAUGAUCUGUC--UGCCCCCCUUGGGCUCUACAUCCCAAUCUCAGUCA--CAGCUUCUCAGAUUCUUCGAGAGAGAG--------- ------------....(((((..(((........(((.(((.--.((((.....))))...))).....))).......--.))).((((.((....)))))))))))--------- ( -19.69, z-score = 0.57, R) >consensus UGCGCUCCCUGAUUCAUUUUUGGGAGUGAUAAAUUCUCACCUGAGGCCCCAGCUUCGAUUUCCAUCUGAAUCUUGGGGC__AAGGCUCUUGAAUCUUCUGAGAAACUG_________ ................((((((((((.((.....))...(((...(((((((....(((((......))))))))))))...))).........))))))))))............. (-12.22 = -13.27 + 1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:17 2011