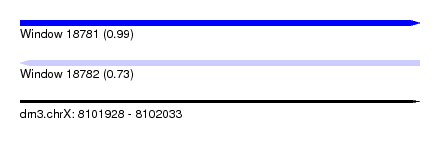
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,101,928 – 8,102,033 |
| Length | 105 |
| Max. P | 0.988765 |

| Location | 8,101,928 – 8,102,033 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.73373 |
| G+C content | 0.45176 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -11.60 |
| Energy contribution | -14.87 |
| Covariance contribution | 3.27 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
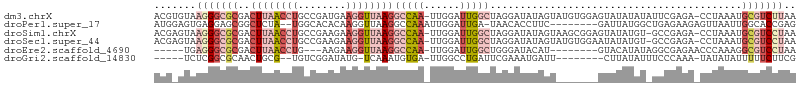
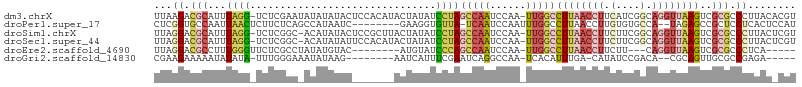
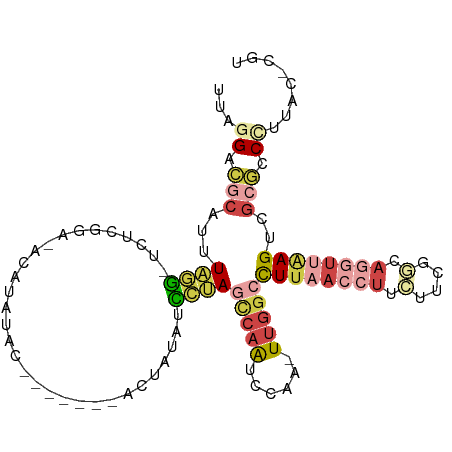
>dm3.chrX 8101928 105 + 22422827 ACGUGUAAGGGCGCGACUUAACCUGCCGAUGAAGGUUAAGGCCAA-UUGGAUUGGCUAGGAUAUAGUAUGUGGAGUAUAUAUAUUCGAGA-CCUAAAUGCGUCUUAA .......((((((((.((((((((........))))))))(((((-(...))))))((((....((((((((.....)))))))).....-))))..)))))))).. ( -32.80, z-score = -2.49, R) >droPer1.super_17 974155 96 + 1930428 AUGGAGUGAGGAGCGGCUCUA--UGGCACACAAGGUUAAGGCCAAAUUGGAUUGA-UAACACCUUC--------GAUUAUGGCUGAGAAGAGUUAAUUGGCACCGAG ...(.(((..((..((((((.--((((.......)))).(((((.....((((((-........))--------)))).)))))....))))))..))..))))... ( -20.90, z-score = 0.28, R) >droSim1.chrX 6457198 104 + 17042790 ACGAGUAAGGGCGCGACUUAACCUGCCGAAGAAGGUUAAGGCCAA-UUGGAUUGGCUAGGAUAUAGUAAGCGGAGUAUAUGU-GCCGAGA-CCUAAAUGCGUCCUAA .......((((((((.((((((((........)))))))).....-..((.(((((..(.(((((.(......).))))).)-)))))..-))....)))))))).. ( -33.10, z-score = -2.00, R) >droSec1.super_44 193211 104 + 251150 ACGAGUAAGGGCGCGACUUAACCUGCCGAAGAAGGUUAAGGCCAA-UUGGAUUGGCUAGGAUAUAGUAUGUGGAAUAUAUGU-GCCGAGA-CCUAAAUGCGUCCUAA .......((((((((.((((((((........))))))))(((((-(...))))))((((.....(((((((.....)))))-)).....-))))..)))))))).. ( -35.80, z-score = -3.39, R) >droEre2.scaffold_4690 10469699 90 + 18748788 -----UGAGGGCGCGACUUAACCUG---AAGAAGGUUAAGGCCAA-UUGGAUUGGCUGGGAUACAU--------GUACAUAUAGGCGAGAACCCAAAGGCGUCCUAA -----..(((((((..((((((((.---....))))))))(((((-(...))))))((((...(.(--------((........))).)..))))...))))))).. ( -33.30, z-score = -3.78, R) >droGri2.scaffold_14830 1678673 89 - 6267026 -----UCUCGGCGCAACUGCG--UGUCGGAUAUG-UCAAAUGUGA-UUGGCCUGAUUCGAAAUGAUU--------CUUAUAUUUCCCAAA-UAUAUAUUUUUCUUCG -----.....((((....)))--).((((((..(-((((......-)))))...)))))).......--------..((((((.....))-))))............ ( -15.00, z-score = 0.25, R) >consensus ACG_GUAAGGGCGCGACUUAACCUGCCGAAGAAGGUUAAGGCCAA_UUGGAUUGGCUAGGAUAUAGU_______GUAUAUAU_GCCGAGA_CCUAAAUGCGUCCUAA .......(((((((..((((((((........))))))))(((((......)))))..........................................))))))).. (-11.60 = -14.87 + 3.27)
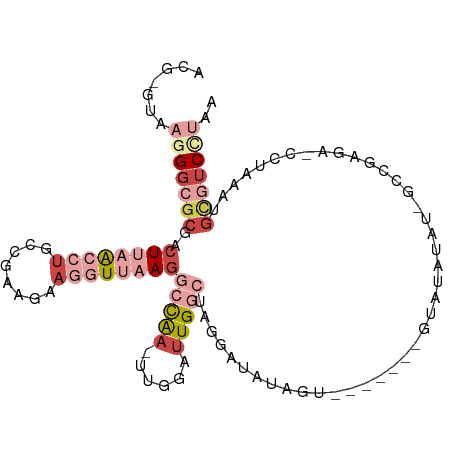
| Location | 8,101,928 – 8,102,033 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.73373 |
| G+C content | 0.45176 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -7.79 |
| Energy contribution | -10.37 |
| Covariance contribution | 2.57 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8101928 105 - 22422827 UUAAGACGCAUUUAGG-UCUCGAAUAUAUAUACUCCACAUACUAUAUCCUAGCCAAUCCAA-UUGGCCUUAACCUUCAUCGGCAGGUUAAGUCGCGCCCUUACACGU .....(((.....(((-.(.(((............................((((((...)-)))))((((((((.(....).))))))))))).).)))....))) ( -21.30, z-score = -1.70, R) >droPer1.super_17 974155 96 - 1930428 CUCGGUGCCAAUUAACUCUUCUCAGCCAUAAUC--------GAAGGUGUUA-UCAAUCCAAUUUGGCCUUAACCUUGUGUGCCA--UAGAGCCGCUCCUCACUCCAU ...((.((.......((((.....(((((((..--------.(((((....-.(((......))))))))....))))).))..--.))))..)).))......... ( -15.20, z-score = 0.34, R) >droSim1.chrX 6457198 104 - 17042790 UUAGGACGCAUUUAGG-UCUCGGC-ACAUAUACUCCGCUUACUAUAUCCUAGCCAAUCCAA-UUGGCCUUAACCUUCUUCGGCAGGUUAAGUCGCGCCCUUACUCGU ..(((.(((...((((-....(((-...........)))........))))((((((...)-)))))((((((((.(....).))))))))..))).)))....... ( -27.90, z-score = -2.25, R) >droSec1.super_44 193211 104 - 251150 UUAGGACGCAUUUAGG-UCUCGGC-ACAUAUAUUCCACAUACUAUAUCCUAGCCAAUCCAA-UUGGCCUUAACCUUCUUCGGCAGGUUAAGUCGCGCCCUUACUCGU ..(((.(((...((((-.......-......................))))((((((...)-)))))((((((((.(....).))))))))..))).)))....... ( -26.05, z-score = -2.35, R) >droEre2.scaffold_4690 10469699 90 - 18748788 UUAGGACGCCUUUGGGUUCUCGCCUAUAUGUAC--------AUGUAUCCCAGCCAAUCCAA-UUGGCCUUAACCUUCUU---CAGGUUAAGUCGCGCCCUCA----- ..(((.(((...((((........(((((....--------))))).))))((((((...)-)))))((((((((....---.))))))))..))).)))..----- ( -24.70, z-score = -2.45, R) >droGri2.scaffold_14830 1678673 89 + 6267026 CGAAGAAAAAUAUAUA-UUUGGGAAAUAUAAG--------AAUCAUUUCGAAUCAGGCCAA-UCACAUUUGA-CAUAUCCGACA--CGCAGUUGCGCCGAGA----- (((((.......((((-((.....))))))..--------.....))))).....((((((-......))).-...........--.((....)))))....----- ( -10.34, z-score = 1.36, R) >consensus UUAGGACGCAUUUAGG_UCUCGGA_ACAUAUAC_______ACUAUAUCCUAGCCAAUCCAA_UUGGCCUUAACCUUCUUCGGCAGGUUAAGUCGCGCCCUUAC_CGU ...((.(((..........................................(((((......)))))((((((((.(....).))))))))..))).))........ ( -7.79 = -10.37 + 2.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:14 2011