
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,082,902 – 8,083,046 |
| Length | 144 |
| Max. P | 0.890252 |

| Location | 8,082,902 – 8,083,046 |
|---|---|
| Length | 144 |
| Sequences | 3 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 75.06 |
| Shannon entropy | 0.32348 |
| G+C content | 0.34871 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -16.98 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
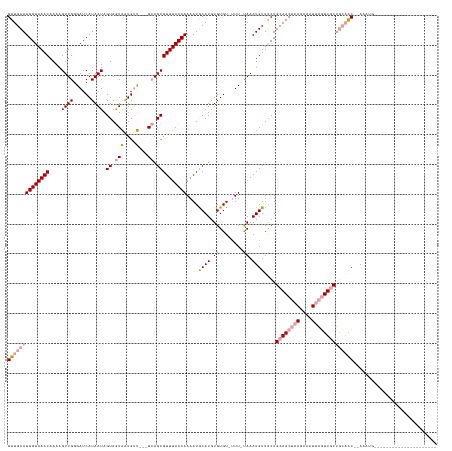
>dm3.chrX 8082902 144 + 22422827 GUUGCAUGGUAAACAAAAGUUGGGAUCAUAAUAGUAAUGGUGACCGAGAAGCGUUUACCAAAAUACAUGUGUGUGCUUUCAAAUGUUGCUGCAACUAAAAAGUACAUUUCGAGUGCCUAAAAGUAAAGCAGCAAACAACAAAUG (((((.((((((((.....((((.(((((.......)))))..)))).....))))))))...(((...((.(..(((..((((((.(((..........))))))))).)))..).))...)))..)))))............ ( -34.20, z-score = -1.20, R) >droSec1.super_44 165980 116 + 251150 GUUGCAUGGUAAACAAAAGUUGGGAUCAUAAUAGUAGUAAUGUU---GCAGCGUUUACCAAAAUGCAUGUGUGU-UUU-UAAAUGUUACUGCAACUAAGAAAUUAGUUGCUGCAAC--AAAUG--------------------- ((((((((((((((....((((.((.(((.((....)).)))))---.))))))))))))((((((....))))-)).-...........((((((((....))))))))))))))--.....--------------------- ( -34.30, z-score = -2.92, R) >droSim1.chrX 6443666 117 + 17042790 GUUGCAUGGUAAACAAAAGUUGGGAUCAUAAUAGUAGUAAUGUU---GCAGCGUUUACCAAAAUGCAUGUGUGU-UUUCUAAAUGUUACUGCAACUAAGAAAUUAGUUGCUGCAAC--AAAUG--------------------- ((((((((((((((....((((.((.(((.((....)).)))))---.))))))))))))((((((....))))-)).............((((((((....))))))))))))))--.....--------------------- ( -34.30, z-score = -2.93, R) >consensus GUUGCAUGGUAAACAAAAGUUGGGAUCAUAAUAGUAGUAAUGUU___GCAGCGUUUACCAAAAUGCAUGUGUGU_UUU_UAAAUGUUACUGCAACUAAGAAAUUAGUUGCUGCAAC__AAAUG_____________________ ((((((((((((((....((((....(((.((....)).)))......))))))))))))....((((.((........)).))))...))))))................................................. (-16.98 = -16.43 + -0.55)


| Location | 8,082,902 – 8,083,046 |
|---|---|
| Length | 144 |
| Sequences | 3 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 75.06 |
| Shannon entropy | 0.32348 |
| G+C content | 0.34871 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.99 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.890252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
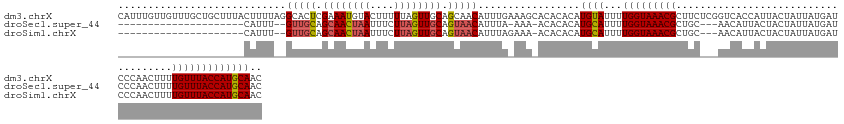
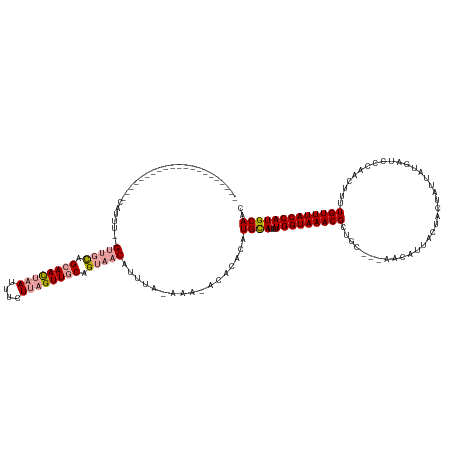
>dm3.chrX 8082902 144 - 22422827 CAUUUGUUGUUUGCUGCUUUACUUUUAGGCACUCGAAAUGUACUUUUUAGUUGCAGCAACAUUUGAAAGCACACACAUGUAUUUUGGUAAACGCUUCUCGGUCACCAUUACUAUUAUGAUCCCAACUUUUGUUUACCAUGCAAC .....(((((.((((((...(((...(((.((.......)).)))...))).))))))((((.((........)).))))....(((((((((......(((((..((....))..)))))........))))))))).))))) ( -29.34, z-score = -1.02, R) >droSec1.super_44 165980 116 - 251150 ---------------------CAUUU--GUUGCAGCAACUAAUUUCUUAGUUGCAGUAACAUUUA-AAA-ACACACAUGCAUUUUGGUAAACGCUGC---AACAUUACUACUAUUAUGAUCCCAACUUUUGUUUACCAUGCAAC ---------------------....(--(((((.((((((((....)))))))).))))))....-...-.......((((...(((((((((....---.............................))))))))))))).. ( -26.21, z-score = -2.82, R) >droSim1.chrX 6443666 117 - 17042790 ---------------------CAUUU--GUUGCAGCAACUAAUUUCUUAGUUGCAGUAACAUUUAGAAA-ACACACAUGCAUUUUGGUAAACGCUGC---AACAUUACUACUAUUAUGAUCCCAACUUUUGUUUACCAUGCAAC ---------------------....(--(((((.((((((((....)))))))).))))))........-.......((((...(((((((((....---.............................))))))))))))).. ( -26.21, z-score = -2.56, R) >consensus _____________________CAUUU__GUUGCAGCAACUAAUUUCUUAGUUGCAGUAACAUUUA_AAA_ACACACAUGCAUUUUGGUAAACGCUGC___AACAUUACUACUAUUAUGAUCCCAACUUUUGUUUACCAUGCAAC ............................(((((.((((((((....)))))))).))))).................((((...(((((((((....................................))))))))))))).. (-17.65 = -18.99 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:09 2011