| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,069,883 – 8,069,988 |
| Length | 105 |
| Max. P | 0.620996 |

| Location | 8,069,883 – 8,069,988 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.64477 |
| G+C content | 0.51437 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
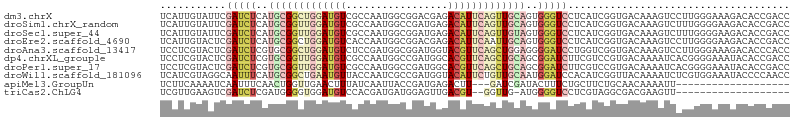
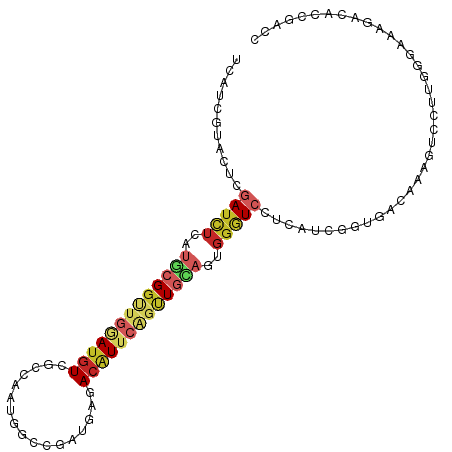
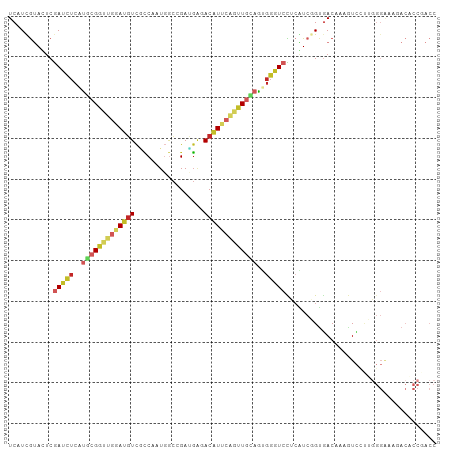
>dm3.chrX 8069883 105 + 22422827 UCAUUGUAUUCGAUCUCAUGCGGCUGGAUGUCGCCAAUGGCGGACGAGACAUUCAGUUGCAGUGGGUCCUCAUCGGUGACAAAGUCCUUGGGAAAGACACCGACC ...........(((((..((((((((((((((.((......))....))))))))))))))..)))))....((((((......(((...)))....)))))).. ( -38.90, z-score = -1.87, R) >droSim1.chrX_random 2428823 105 + 5698898 UCAUUGUAUUCGAUCUCAUGCGGUUGGAUGUCGCCAAUGGCCGAUGAGACAUUCAGUUGCAGUGGGUCCUCAUCGGUGACAAAGUCUUUGGGGAAGACACCGACC .....((((........))))((((((.((((.((((..(((((((((..(((((.......))))).)))))))))(((...))).))))....)))))))))) ( -33.30, z-score = -0.34, R) >droSec1.super_44 156130 105 + 251150 UCAUUGUAUUCGAUCUCAUGCGGUUGGAUGUCGCCAAUGGCGGAUGAGACAUUCAGUUGUAGUGGGUCCUCAUCGGUGACAAAGUCUUUGGGGAAGACACCGACC .....((((........))))((((((.(((((((.((((.((((...(((......))).....)))))))).)))))))..((((((...)))))).)))))) ( -34.10, z-score = -0.78, R) >droEre2.scaffold_4690 10441190 105 + 18748788 UCAUUGUACUCGAUCUCAUGCGGCUGGAUGUCACCAAUGGCGGACGAGACAUUCAAUUGCAGUGGGUCCUCAUCGGUGACAAAGUCCUUGGGGAAGACACCGACC ...........(((((..(((((.((((((((.((......))....)))))))).)))))..)))))....((((((......(((....)))...)))))).. ( -31.90, z-score = 0.09, R) >droAna3.scaffold_13417 407403 105 + 6960332 UCCUCGUACUCGAUCUCGUGCGGCUGGAUGUCUCCGAUGGCGGAUGGUACGUUCAGCUGGAGGGGAUCCUGGUCGGUGACAAAGUCCUUGGGAAAGACACCCACC ...........((((((.(.((((((((((((((((....))))..).))))))))))).).))))))..(((.((((......(((...)))....)))).))) ( -39.50, z-score = -0.46, R) >dp4.chrXL_group1e 4002242 105 + 12523060 UCCUCGUACUCGAUCUCGUGCGGUUGGAUGUCGCCAAUGGCCGAUGGCACGUUCAGCUGCAGCGGAUCUUCGUCCGUGACAAAAUCACGGGGAAAUACACCGACC .....(((...((((.(((((((((((((((.((((........))))))))))))))))).))))))(((.(((((((.....)))))))))).)))....... ( -43.10, z-score = -2.84, R) >droPer1.super_17 937205 105 + 1930428 UCCUCGUACUCGAUCUCGUGCGGUUGGAUGUCGCCAAUGGCCGAUGGCACGUUCAGCUGCAGCGGAUCUUCGUCCGUGACAAAAUCACGGGGAAAUACACCGACC .....(((...((((.(((((((((((((((.((((........))))))))))))))))).))))))(((.(((((((.....)))))))))).)))....... ( -43.10, z-score = -2.84, R) >droWil1.scaffold_181096 5553734 105 - 12416693 UCAUCGUAGGCAAUUUCAUGCGGCUGAAUGUUACCAAUCGCCGAUGGUACAUUCUGUUGCAAUGGAUCCACAUCGGUUACAAAAUCUCGUGGAAAUACCCCAACC .....((.((....((((((((((.((((((.((((........)))))))))).))))).)))))(((((...((((....))))..)))))......)).)). ( -29.10, z-score = -1.69, R) >apiMel3.GroupUn 29092764 83 + 399230636 UCUUCAAAAUCAAUUUCAACUGGUUGAACUUUAUCAAUUACCGAUGAGACUU---GAUCGAUACUUUCUGCUUCUGCAACAAAAUU------------------- ........((((((((((.((((((((......)))))))..).))))).))---)))..........(((....)))........------------------- ( -11.20, z-score = -0.97, R) >triCas2.ChLG4 7234289 83 + 13894384 UCGUUGAAGUCGAUCUCGAUGGGGUGGAUGUCCACGAUGAUGGAGUUGACGU--GGUUG-AUGGGGUCCUCGUAGGCGACGAAGUU------------------- (((((...(((((.(((.((.(.((((....))))..).)).))))))))..--.((..-(((((...)))))..)))))))....------------------- ( -25.80, z-score = -0.44, R) >consensus UCAUCGUACUCGAUCUCAUGCGGUUGGAUGUCGCCAAUGGCCGAUGAGACAUUCAGUUGCAGUGGGUCCUCAUCGGUGACAAAGUCCUUGGGAAAGACACCGACC ...........(((((..(((((((((((((.................)))))))))))))..)))))..................................... (-13.17 = -14.22 + 1.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:07 2011