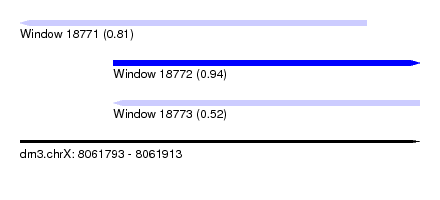
| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,061,793 – 8,061,913 |
| Length | 120 |
| Max. P | 0.936486 |

| Location | 8,061,793 – 8,061,897 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.61891 |
| G+C content | 0.59805 |
| Mean single sequence MFE | -37.31 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.15 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8061793 104 - 22422827 UCAGCAGCGAGUACCAUAUCGCUGGUACUCGGGAGCUCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGAAAGAGCAGGACAGGCACAUCCAGGCAAG---- ...(((((((........)))))((((((((..((..((.((((....)))).))..)).))))))))((((((...........))))))........))...---- ( -36.60, z-score = -1.23, R) >droEre2.scaffold_4690 10433242 104 - 18748788 CCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGAGCGAGCAGGACAGGCUCAUCUAGGCGUG---- .......((((((((((((.....)))...))))))))).((((....))))((((.((((((((.(((((((....))))).))....)))))..))))))).---- ( -41.60, z-score = -0.38, R) >droYak2.chrX 8522336 104 + 21770863 CCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGUGCACGGCGUUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGAAAGAGCAGGACAGGCUCAUCUAGGCAAG---- .....(((((........)))))(((((((((((((.(((((....)).)))..))))).))))))))(((((.((..((((.......)))).)))))))...---- ( -38.60, z-score = -0.93, R) >droSec1.super_44 147930 104 - 251150 UCAGCAGCGGGUACCAUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGAAAGAGCAGGACAGGCUCAACCAGGCAAG---- ...((..(((((((((......)))))))))((.((((((((....)).)))).))....(((((....(((((......)))))....))))).))..))...---- ( -39.10, z-score = -1.51, R) >droSim1.chrX_random 2419254 104 - 5698898 UCAGCAGCGGGUACCCUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGAAAGAGCAGGACAGGCUCAACCAGGCAAG---- .....(((((........)))))((((((((((.((((((((....)).)))).)).)).))))))))(((((.(...((((.......))))..))))))...---- ( -38.70, z-score = -0.97, R) >droAna3.scaffold_13417 398180 96 - 6960332 UCAGCAGCGGGUUCCGUAUCGCUGGUAUUCCGGAGCUCGCCGGUCACGGUCGGCGCAUUACGAGUACCGCCUGUGAGUGAAAGAGAGGGAAGCCGA------------ ......(((((((((((((.....)))...))))))))))((.((((((.(((..(.......)..))).)))))).)).......((....))..------------ ( -35.40, z-score = -0.20, R) >droVir3.scaffold_12970 8378586 83 - 11907090 UCAGCAGCGGGUGCCAUAUCGCUGGUACUCGGGUGCGCGGCGUUCACGUUCCGCGCACUACGAGUACCGUCUCUGAAAGAAAC------------------------- ((((.(((((........)))))(((((((((((((((((((....))..))))))))).))))))))....)))).......------------------------- ( -40.50, z-score = -4.19, R) >droMoj3.scaffold_6482 1840146 92 + 2735782 CCAGCAGCGGGUGCCGUAUCGCUGGUAUUCCGGUGCGCGGCGUUCUCGCUCCGCUCACUACGAAUAUCGACUCUGAGAG--CCGGCAAGAGACA-------------- ((.((((((((((((((..((((((....)))))).))))))..))))))...((((...((.....))....)))).)--).)).........-------------- ( -32.10, z-score = 0.35, R) >droGri2.scaffold_14853 1915887 99 + 10151454 UCAGCAGCGUGUGCCGUAUCGUUGGUAUUCCGGUGCGAGACGUUCACGUUCCGCUCACUAUGAAUACCGCCUCUGAGAG----AGACAGCAAUAGCGAGACAA----- ......((...(((.((......(((((((.((((((..(((....)))..)))..)))..)))))))..(((.....)----)))).)))...)).......----- ( -28.70, z-score = -0.19, R) >droWil1.scaffold_181096 5544907 87 + 12416693 UCAGCAACGUGUCCCCUAUCGUUGGUAUUCAGGUGCACGACGUUCUCGUUCCGCACACUAUGAAUAUCGUUUGUGAGUG----GGUGCGAG----------------- .......((..(((.((.....(((((((((.((((..((((....))))..))))....)))))))))......)).)----))..))..----------------- ( -28.60, z-score = -1.64, R) >droPer1.super_17 928114 108 - 1930428 CCAGCAACGUGUUCCCUAUCGCUGGUACUCGGGUGCUCGACGCUCCCGCUCGGCCCACUACGAGUACCGGCUGUGAGAGAGCGGGACACCAGCAGGAAAGGGACAGCA ........((..(((((.(((((((((((((((((.((((.((....))))))..)))).))))))))))).........((((....)).))..)).)))))..)). ( -43.90, z-score = -0.93, R) >dp4.chrXL_group1e 3993036 108 - 12523060 CCAGCAACGUGUUCCCUAUCGCUGGUACUCGGGUGCUCGACGCUCCCGCUCGGCCCACUACGAGUACCGGCUGUGAGAGAGCGGGACACCAGCAGGAAAGGGACAGCA ........((..(((((.(((((((((((((((((.((((.((....))))))..)))).))))))))))).........((((....)).))..)).)))))..)). ( -43.90, z-score = -0.93, R) >consensus UCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGUGCUCGGCGUUCUCGAUCGGCGCACUAUGAGUACCGCCUGUGAGAGAGCAGGACAGGCUCAGC_AGGCAA_____ ...((((((..........))))((((((((.(.(((.((((....)).))))).)....))))))))))...................................... (-18.80 = -18.15 + -0.65)
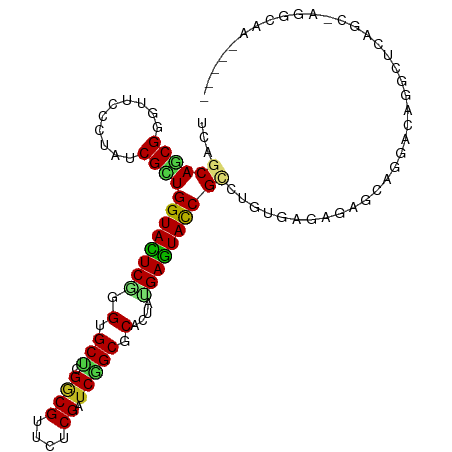
| Location | 8,061,821 – 8,061,913 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.52322 |
| G+C content | 0.61299 |
| Mean single sequence MFE | -37.81 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8061821 92 + 22422827 ----UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAUCGCCGAGCUCCCGAGUACCAGCGAUAUGGUACUCGCUGCUGAUGCAGCGCCAAGUCAG------------------------ ----......(((((((((..(((.((.((((....)))).)).)))...))))))).))(((.(((....((((((....))))))))).)))..------------------------ ( -37.10, z-score = -2.83, R) >droEre2.scaffold_4690 10433270 92 + 18748788 ----UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAUCGCCGGGCUCCCGAGUACCAGCGAUAGGGAACCCGCUGCUGGUGCAGCGCCAGGCCAG------------------------ ----......(((((((((..(((.((.((((....)))).)).)))...))))))).((....((....))(((((....)))))))...))...------------------------ ( -36.80, z-score = -1.05, R) >droYak2.chrX 8522364 92 - 21770863 ----UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAACGCCGUGCACCCGAGUACCAGCGAUAGGGAACCCGCUGCUGGUGCAGCGCCAGGCCAG------------------------ ----......(((((((((...((((((((........))..))))))..))))))).((....((....))(((((....)))))))...))...------------------------ ( -35.10, z-score = -1.33, R) >droSec1.super_44 147958 92 + 251150 ----UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAUCGCCGGGCUCCCGAGUACCAGCGAUAUGGUACCCGCUGCUGAUGCAGCGCCAAGCCAG------------------------ ----......(((((((((..(((.((.((((....)))).)).)))...))))))).))....((((...((((((....))))))....)))).------------------------ ( -38.40, z-score = -2.83, R) >droSim1.chrX_random 2419282 92 + 5698898 ----UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAUCGCCGGGCUCCCGAGUACCAGCGAUAGGGUACCCGCUGCUGAUGCAGCGCCAAGCCAG------------------------ ----......(((((((((..(((.((.((((....)))).)).)))...))))))).)).....(((...((((((....))))))....)))..------------------------ ( -37.40, z-score = -2.15, R) >droAna3.scaffold_13417 398200 116 + 6960332 ----UCACAGGCGGUACUCGUAAUGCGCCGACCGUGACCGGCGAGCUCCGGAAUACCAGCGAUACGGAACCCGCUGCUGAUGAAGUGCCAGUCCUGAUCCUGUUCCAGCUCCUGAUGCAG ----...((((((((((((((....(((((........)))))(((..(((....((........))...)))..))).))).)))))).).))))...((((..(((...)))..)))) ( -37.00, z-score = -0.42, R) >droVir3.scaffold_12970 8378593 104 + 11907090 ----UCAGAGACGGUACUCGUAGUGCGCGGAACGUGAACGCCGCGCACCCGAGUACCAGCGAUAUGGCACCCGCUGCUGAUGCAACGCCAGGCCCAGAGCCAGACCGG------------ ----........((((((((..((((((((..((....)))))))))).))))))))..((...((((....(.(((....))).)))))(((.....)))....)).------------ ( -43.30, z-score = -2.65, R) >droMoj3.scaffold_6482 1840158 108 - 2735782 GCUCUCAGAGUCGAUAUUCGUAGUGAGCGGAGCGAGAACGCCGCGCACCGGAAUACCAGCGAUACGGCACCCGCUGCUGGUGCAAGGCCAGGCCCAGGGCCAAGCCAG------------ ((((.(.((((....))))...).))))...(((.......)))(((((((.....(((((..........))))))))))))..(((..((((...))))..)))..------------ ( -39.60, z-score = -0.22, R) >droGri2.scaffold_14853 1915910 104 - 10151454 ----UCAGAGGCGGUAUUCAUAGUGAGCGGAACGUGAACGUCUCGCACCGGAAUACCAACGAUACGGCACACGCUGCUGAUGCAGCGCAACACCCAAUGCCAGAUUGG------------ ----.....((((((((((...(.(.(((..(((....)))..))).)).)))))))........((....((((((....))))))......))...))).......------------ ( -38.40, z-score = -2.85, R) >droWil1.scaffold_181096 5544918 99 - 12416693 ----UCACAAACGAUAUUCAUAGUGUGCGGAACGAGAACGUCGUGCACCUGAAUACCAACGAUAGGGGACACGUUGCUGAUGCAAUCCCAGACCCAAACCCAA----------------- ----........(.((((((..(((..((..(((....)))))..))).)))))).).......(((.....(((((....)))))......)))........----------------- ( -23.80, z-score = -0.88, R) >droPer1.super_17 928146 101 + 1930428 ----UCACAGCCGGUACUCGUAGUGGGCCGAGCGGGAGCGUCGAGCACCCGAGUACCAGCGAUAGGGAACACGUUGCUGGUGCAGCGAUAAGCCCAUGCUGCCAG--------------- ----...((((.((((((((....(((((((((....)).))).)).)))))))))).......(((....((((((....)))))).....)))..))))....--------------- ( -43.40, z-score = -2.10, R) >dp4.chrXL_group1e 3993068 101 + 12523060 ----UCACAGCCGGUACUCGUAGUGGGCCGAGCGGGAGCGUCGAGCACCCGAGUACCAGCGAUAGGGAACACGUUGCUGGUGCAACGACAAGCCCAUGCUGCCAG--------------- ----...((((.((((((((....(((((((((....)).))).)).)))))))))).......(((....((((((....)))))).....)))..))))....--------------- ( -43.40, z-score = -2.25, R) >consensus ____UCACAAGCGGUACUCAUAGUGCGCCGAUCGAGAACGCCGAGCACCCGAGUACCAGCGAUAGGGAACCCGCUGCUGAUGCAGCGCCAGGCCCA________________________ ............(((((((...(.(.((....((....))....)).)).)))))))........((....((((((....)))))).....)).......................... (-19.22 = -19.43 + 0.21)


| Location | 8,061,821 – 8,061,913 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.52322 |
| G+C content | 0.61299 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -21.28 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 8061821 92 - 22422827 ------------------------CUGACUUGGCGCUGCAUCAGCAGCGAGUACCAUAUCGCUGGUACUCGGGAGCUCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGA---- ------------------------..((..(((((((((....))))))....)))..))((.((((((((..((..((.((((....)))).))..)).))))))))))......---- ( -37.40, z-score = -1.60, R) >droEre2.scaffold_4690 10433270 92 - 18748788 ------------------------CUGGCCUGGCGCUGCACCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGA---- ------------------------..(((..((((((((....)))))((....))....)))((((((((((.((((((((....)).)))).)).)).))))))))))).....---- ( -38.80, z-score = -0.64, R) >droYak2.chrX 8522364 92 + 21770863 ------------------------CUGGCCUGGCGCUGCACCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGUGCACGGCGUUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGA---- ------------------------..(((..((((((((....)))))((....))....)))(((((((((((((.(((((....)).)))..))))).))))))))))).....---- ( -39.50, z-score = -0.98, R) >droSec1.super_44 147958 92 - 251150 ------------------------CUGGCUUGGCGCUGCAUCAGCAGCGGGUACCAUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGA---- ------------------------...(((.((((((((....)))))((((((((......))))))))....))).)))..((.(((.(((..(.......)..))).))).))---- ( -37.90, z-score = -0.92, R) >droSim1.chrX_random 2419282 92 - 5698898 ------------------------CUGGCUUGGCGCUGCAUCAGCAGCGGGUACCCUAUCGCUGGUACUCGGGAGCCCGGCGAUCUCGAUCGGCGCACUAUGAGUACCGCUUGUGA---- ------------------------..(((..((((((((....)))))((....))....)))((((((((((.((((((((....)).)))).)).)).))))))))))).....---- ( -39.20, z-score = -1.10, R) >droAna3.scaffold_13417 398200 116 - 6960332 CUGCAUCAGGAGCUGGAACAGGAUCAGGACUGGCACUUCAUCAGCAGCGGGUUCCGUAUCGCUGGUAUUCCGGAGCUCGCCGGUCACGGUCGGCGCAUUACGAGUACCGCCUGUGA---- ...((.((((.(((((...((..(((....)))..))...)))))(((((........)))))(((((((..(.(((.((((....)))).))).).....))))))).)))))).---- ( -39.00, z-score = 0.73, R) >droVir3.scaffold_12970 8378593 104 - 11907090 ------------CCGGUCUGGCUCUGGGCCUGGCGUUGCAUCAGCAGCGGGUGCCAUAUCGCUGGUACUCGGGUGCGCGGCGUUCACGUUCCGCGCACUACGAGUACCGUCUCUGA---- ------------.((((.((((.....(((...((((((....))))))))))))).))))..(((((((((((((((((((....))..))))))))).))))))))........---- ( -50.60, z-score = -2.37, R) >droMoj3.scaffold_6482 1840158 108 + 2735782 ------------CUGGCUUGGCCCUGGGCCUGGCCUUGCACCAGCAGCGGGUGCCGUAUCGCUGGUAUUCCGGUGCGCGGCGUUCUCGCUCCGCUCACUACGAAUAUCGACUCUGAGAGC ------------..((((.((((...)))).)))).(((....)))(((((((((((..((((((....)))))).))))))..)))))...((((.(..((.....)).....).)))) ( -42.90, z-score = -0.09, R) >droGri2.scaffold_14853 1915910 104 + 10151454 ------------CCAAUCUGGCAUUGGGUGUUGCGCUGCAUCAGCAGCGUGUGCCGUAUCGUUGGUAUUCCGGUGCGAGACGUUCACGUUCCGCUCACUAUGAAUACCGCCUCUGA---- ------------.......(((....((..(.(((((((....))))))))..))........(((((((.((((((..(((....)))..)))..)))..)))))))))).....---- ( -40.10, z-score = -2.09, R) >droWil1.scaffold_181096 5544918 99 + 12416693 -----------------UUGGGUUUGGGUCUGGGAUUGCAUCAGCAACGUGUCCCCUAUCGUUGGUAUUCAGGUGCACGACGUUCUCGUUCCGCACACUAUGAAUAUCGUUUGUGA---- -----------------.........(((..((((((((....)))....)))))..)))...((((((((.((((..((((....))))..))))....))))))))........---- ( -27.70, z-score = -0.03, R) >droPer1.super_17 928146 101 - 1930428 ---------------CUGGCAGCAUGGGCUUAUCGCUGCACCAGCAACGUGUUCCCUAUCGCUGGUACUCGGGUGCUCGACGCUCCCGCUCGGCCCACUACGAGUACCGGCUGUGA---- ---------------..((.(((((((((.....)))((....))..)))))).))(((.(((((((((((((((.((((.((....))))))..)))).))))))))))).))).---- ( -40.60, z-score = -1.20, R) >dp4.chrXL_group1e 3993068 101 - 12523060 ---------------CUGGCAGCAUGGGCUUGUCGUUGCACCAGCAACGUGUUCCCUAUCGCUGGUACUCGGGUGCUCGACGCUCCCGCUCGGCCCACUACGAGUACCGGCUGUGA---- ---------------...((((.(((((.....((((((....)))))).....))))))(((((((((((((((.((((.((....))))))..)))).))))))))))))))..---- ( -43.00, z-score = -1.65, R) >consensus ________________________CGGGCCUGGCGCUGCAUCAGCAGCGGGUUCCCUAUCGCUGGUACUCGGGUGCUCGGCGUUCUCGAUCGGCGCACUAUGAGUACCGCCUGUGA____ ...........................(((((...((((....)))))))))...........((((((((.(.(((.((((....)).))))).)....))))))))............ (-21.28 = -20.37 + -0.91)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:06 2011