| Sequence ID | dm3.chrX |
|---|---|
| Location | 8,044,152 – 8,044,221 |
| Length | 69 |
| Max. P | 0.712981 |

| Location | 8,044,152 – 8,044,221 |
|---|---|
| Length | 69 |
| Sequences | 12 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.62091 |
| G+C content | 0.46638 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.02 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
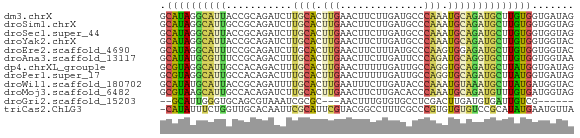
>dm3.chrX 8044152 69 - 22422827 GCAUAGGCAUUACCGCAGAUCUUGCACUUGAACUUCUUGAUGCCCAAAUGCAGAUGCUUGUGGUGAUAG ((....))((((((((((((((.(((.(((..(........)..))).))))))).).))))))))).. ( -19.40, z-score = -1.38, R) >droSim1.chrX 6433722 69 - 17042790 GCAUAGGCAUUGCCGCAGAUCUUGCACUUGAACUUCUUGAUGCCCAAAUGCAGAUGCUUGUGGUGGUAG ((....))((..((((((((((.(((.(((..(........)..))).))))))).).)))))..)).. ( -19.10, z-score = -0.13, R) >droSec1.super_44 130618 69 - 251150 GCAUAGGCAUUACCGCAGAUCUUGCACUUGAACUUCUUGAUGCCCAAAUGCAGAUGCUUGUGGUGGUAG ((....))((((((((((((((.(((.(((..(........)..))).))))))).).))))))))).. ( -18.90, z-score = -0.64, R) >droYak2.chrX 8504795 69 + 21770863 GCAUAGGCAUUACCGCAGAUCUUGCACUUGAACUUCUUGAUGCCCAAAUGCAGAUGCUUGUGGUGGUAC ((....))((((((((((((((.(((.(((..(........)..))).))))))).).))))))))).. ( -18.90, z-score = -0.82, R) >droEre2.scaffold_4690 10415755 69 - 18748788 GCAUAGGCAUUUCCGCAGAUCUUGCACUUGAACUUCUUUAUGCCCAAGUGGAGAUGCUUGUGGUGGUAC .(((((((((((((((((...))))(((((..(........)..))))))))))))))))))....... ( -25.20, z-score = -2.94, R) >droAna3.scaffold_13117 2247070 69 - 5790199 GCAUAUGCGUUUCCGCAGACUUUGCACUUGAACUUCUUGAUUCCCAGAUGCAGGUGCUUGUGGUGGUAA ((....))....((((..((...(((((((....(((.(....).)))..)))))))..)).))))... ( -18.80, z-score = -0.69, R) >dp4.chrXL_group1e 3974128 69 - 12523060 GCGUAGGCAUUGCCACAGACUUUGCACUUGAACUUUUUGAUUGCCAGGUGCAGAUGCUUAUGGUGAUAG ((....))((..(((.((..((((((((((..............))))))))))..))..)))..)).. ( -19.84, z-score = -1.31, R) >droPer1.super_17 909154 69 - 1930428 GCGUAGGCAUUGCCACAGACUUUGCACUUGAACUUUUUGAUUGCCAGGUGCAGAUGCUUAUGGUGAUAG ((....))((..(((.((..((((((((((..............))))))))))..))..)))..)).. ( -19.84, z-score = -1.31, R) >droWil1.scaffold_180702 632047 69 - 4511350 GCAUAUGCAUUACCGCAGAUUUUGCACUUGAAUUUCUUGAUACCCAAAUGUAAAUGCUUAUGAUGGUAC ((....))..(((((((...((((((.(((..............))).))))))......)).))))). ( -10.34, z-score = -0.03, R) >droMoj3.scaffold_6482 1820176 69 + 2735782 GCGUAAGCAUUGCCACAGAUCUUGCACUUGAACUUCUUGACACCCAAAUGCAGAUGUUUGUGAUGGUAG ((....))(((..(((((((.(((((.(((..............))).)))))..)))))))..))).. ( -15.94, z-score = -0.61, R) >droGri2.scaffold_15203 3464128 58 + 11997470 --GCAUUGGGUGCAGCGUAAAUCGCGC---AACUUUGUGUGCCUCGACUUGAUGUGAUUGUCG------ --((((.((((((.(((.....)))))---.))))...))))..((((...........))))------ ( -14.30, z-score = 0.38, R) >triCas2.ChLG3 27817928 68 - 32080666 -CAUAUUUCUGGUUGCACAAUUCGCAUUCGUACGGCCUUUCGCCCGUGUGUGUCCGCAUAUGAAUGUUA -......................((((((((..(((.....))).(((((....))))))))))))).. ( -13.90, z-score = 0.12, R) >consensus GCAUAGGCAUUGCCGCAGAUCUUGCACUUGAACUUCUUGAUGCCCAAAUGCAGAUGCUUGUGGUGGUAG .((((((((((...........((((.(((..............))).))))))))))))))....... ( -9.46 = -9.02 + -0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:25:02 2011